2VLK
 
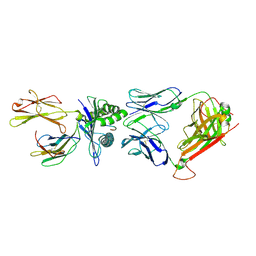 | | The Structural Dynamics and Energetics of an Immunodominant T-cell Receptor are Programmed by its Vbeta Domain | | Descriptor: | BETA-2-MICROGLOBULIN, FLU MATRIX PEPTIDE, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Ishizuka, J, Stewart-Jones, G, Van Der Merwe, A, Bell, J, Mcmichael, A, Jones, Y. | | Deposit date: | 2008-01-15 | | Release date: | 2008-01-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Structural Dynamics and Energetics of an Immunodominant T-Cell Receptor are Programmed by its Vbeta Domain
Immunity, 28, 2008
|
|
4YYE
 
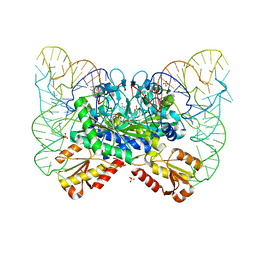 | |
6TPP
 
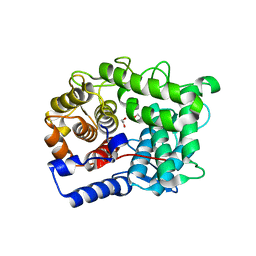 | |
7BYW
 
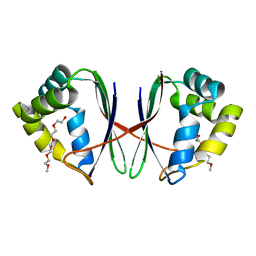 | |
2C36
 
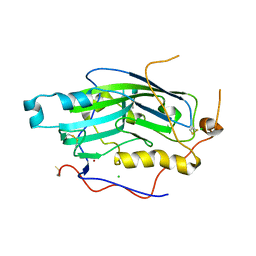 | | Structure of unliganded HSV gD reveals a mechanism for receptor- mediated activation of virus entry | | Descriptor: | CHLORIDE ION, GLYCOPROTEIN D HSV-1, ZINC ION, ... | | Authors: | Krummenacher, C, Supekar, V.M, Whitbeck, J.C, Lazear, E, Connolly, S.A, Eisenberg, R.J, Cohen, G.H, Wiley, D.C, Carfi, A. | | Deposit date: | 2005-10-04 | | Release date: | 2005-11-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure of unliganded HSV gD reveals a mechanism for receptor-mediated activation of virus entry.
EMBO J., 24, 2005
|
|
4Z24
 
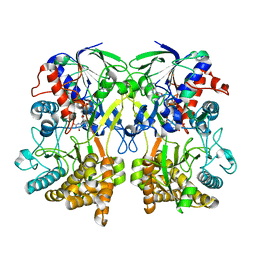 | | Mimivirus R135 (residues 51-702) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GMC-type oxidoreductase R135 | | Authors: | Klose, T, Rossmann, M.G. | | Deposit date: | 2015-03-28 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Mimivirus Enzyme that Participates in Viral Entry.
Structure, 23, 2015
|
|
4Z2T
 
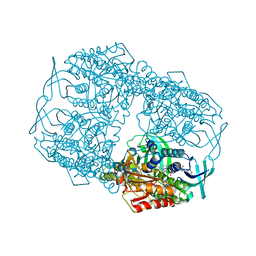 | |
2NO4
 
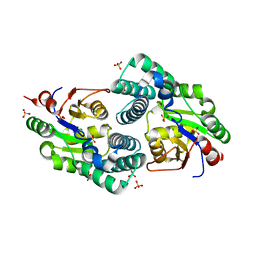 | | Crystal Structure analysis of a Dehalogenase | | Descriptor: | (S)-2-haloacid dehalogenase IVA, CHLORIDE ION, SULFATE ION | | Authors: | Schmidberger, J.W, Wilce, M.C.J. | | Deposit date: | 2006-10-24 | | Release date: | 2007-09-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of the substrate free-enzyme, and reaction intermediate of the HAD superfamily member, haloacid dehalogenase DehIVa from Burkholderia cepacia MBA4
J.Mol.Biol., 368, 2007
|
|
2NC9
 
 | | Apo solution structure of Hop TPR2A | | Descriptor: | Stress-induced-phosphoprotein 1 | | Authors: | Darby, J.F, Vidler, L.R, Simpson, P.J, Matthews, S.J, Sharp, S.Y, Pearl, L.H, Hoelder, S, Workman, P. | | Deposit date: | 2016-03-23 | | Release date: | 2017-03-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Hop TPR2A domain and investigation of target druggability by NMR, biochemical and in silico approaches.
Sci Rep, 10, 2020
|
|
2BXN
 
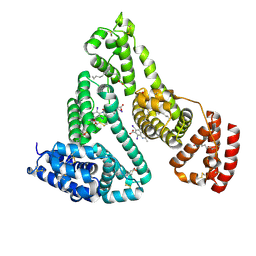 | | Human serum albumin complexed with myristate and iodipamide | | Descriptor: | 3-[5-[(3-CARBOXY-2,4,6-TRIIODO-PHENYL)CARBAMOYL]PENTANOYLAMINO]-2,4,6-TRIIODO-BENZOIC ACID, MYRISTIC ACID, SERUM ALBUMIN | | Authors: | Ghuman, J, Zunszain, P.A, Petitpas, I, Bhattacharya, A.A, Curry, S. | | Deposit date: | 2005-07-26 | | Release date: | 2005-09-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural Basis of the Drug-Binding Specificity of Human Serum Albumin.
J.Mol.Biol., 353, 2005
|
|
4Z5Q
 
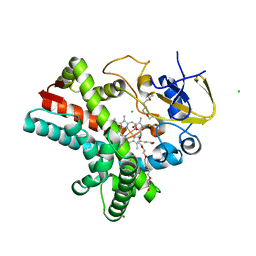 | | Crystal structure of the LnmZ cytochrome P450 hydroxylase from the leinamycin biosynthetic pathway of Streptomyces atroolivaceus S-140 at 1.8 A resolution | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, Cytochrome P450 hydroxylase, ... | | Authors: | Ma, M, Lohman, J, Rudolf, J, Miller, M.D, Cao, H, Osipiuk, J, Joachimiak, A, Phillips Jr, G.N, Shen, B, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-04-02 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal structure of the LnmZ cytochrome P450 hydroxylase from the leinamycin biosynthetic pathway of Streptomyces atroolivaceus S-140
To be Published
|
|
4C43
 
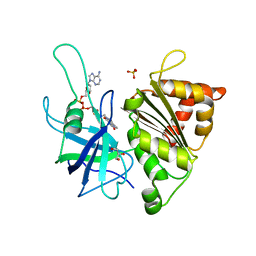 | | FERREDOXIN NADP REDUCTASE MUTANT WITH GLU 103 REPLACED BY TYR, TYR 104 REPLACED BY PHE, SER 109 REPLACED BY PHE AND GLY 110 REPLACED BY PRO (E103Y-Y104F-S109F-G110P) | | Descriptor: | FERREDOXIN--NADP REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Martinez-Julvez, M, Herguedas, B, Sanchez-Azqueta, A, Hervas, M, Navarro, J.A, Medina, M. | | Deposit date: | 2013-08-29 | | Release date: | 2013-12-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | External Loops at the Ferredoxin-Nadp(+) Reductase Protein-Partner Binding Cavity Contribute to Substrates Allocation.
Biochim.Biophys.Acta, 1837, 2013
|
|
4ZE2
 
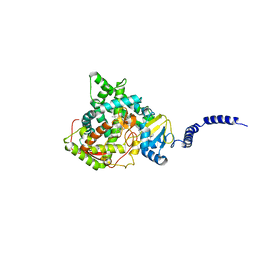 | | Saccharomyces cerevisiae CYP51 (Lanosterol 14-alpha demethylase) Y140H mutant complexed with itraconazole | | Descriptor: | 2-[(2R)-butan-2-yl]-4-{4-[4-(4-{[(2R,4S)-2-(2,4-dichlorophenyl)-2-(1H-1,2,4-triazol-1-ylmethyl)-1,3-dioxolan-4-yl]methoxy}phenyl)piperazin-1-yl]phenyl}-2,4-dihydro-3H-1,2,4-triazol-3-one, Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sagatova, A, Keniya, M.V, Wilson, R, Monk, B.C, Tyndall, J.D.A. | | Deposit date: | 2015-04-20 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Triazole resistance mediated by mutations of a conserved active site tyrosine in fungal lanosterol 14 alpha-demethylase.
Sci Rep, 6, 2016
|
|
2BV2
 
 | | beta gamma crystallin from Ciona Intestinalis | | Descriptor: | ACETATE ION, CALCIUM ION, CIONA BETAGAMMA-CRYSTALLIN, ... | | Authors: | Shimeld, S.M, Purkiss, A.G, Dirks, R.P.H, Bateman, O.A, Slingsby, C, Lubsen, N.H. | | Deposit date: | 2005-06-21 | | Release date: | 2005-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Urochordate Betagamma-Crystallin and the Evolutionary Origin of the Vertebrate Eye Lens.
Curr.Biol., 15, 2005
|
|
2BXE
 
 | | Human serum albumin complexed with diflunisal | | Descriptor: | 5-(2,4-DIFLUOROPHENYL)-2-HYDROXY-BENZOIC ACID, SERUM ALBUMIN | | Authors: | Ghuman, J, Zunszain, P.A, Petitpas, I, Bhattacharya, A.A, Curry, S. | | Deposit date: | 2005-07-26 | | Release date: | 2005-09-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Basis of the Drug-Binding Specificity of Human Serum Albumin.
J.Mol.Biol., 353, 2005
|
|
2I2Z
 
 | | Human serum albumin complexed with myristate and aspirin | | Descriptor: | 2-HYDROXYBENZOIC ACID, MYRISTIC ACID, Serum albumin | | Authors: | Yang, F, Bian, C, Zhu, L, Zhao, G, Huang, Z, Huang, M. | | Deposit date: | 2006-08-17 | | Release date: | 2006-12-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Effect of human serum albumin on drug metabolism: Structural evidence of esterase activity of human serum albumin
J.Struct.Biol., 157, 2007
|
|
2VLR
 
 | | The Structural Dynamics and Energetics of an Immunodominant T-cell Receptor are Programmed by its Vbeta Domain | | Descriptor: | BETA-2-MICROGLOBULIN, FLU MATRIX PEPTIDE, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Ishizuka, J, Stewart-Jones, G, van der Merwe, A, Bell, J, McMichael, A, Jones, Y. | | Deposit date: | 2008-01-15 | | Release date: | 2008-01-22 | | Last modified: | 2019-09-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structural Dynamics and Energetics of an Immunodominant T-Cell Receptor are Programmed by its Vbeta Domain
Immunity, 28, 2008
|
|
7BVQ
 
 | | Structure of human beta1 adrenergic receptor bound to carazolol | | Descriptor: | (2S)-1-(9H-Carbazol-4-yloxy)-3-(isopropylamino)propan-2-ol, (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, CHOLESTEROL, ... | | Authors: | Xu, X, Kaindl, J, Clark, M, Hubner, H, Hirata, K, Sunahara, R, Gmeiner, P, Kobilka, B.K, Liu, X. | | Deposit date: | 2020-04-11 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Binding pathway determines norepinephrine selectivity for the human beta 1 AR over beta 2 AR.
Cell Res., 31, 2021
|
|
2Y76
 
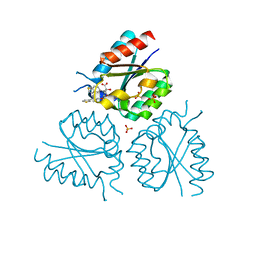 | | Structure of Mycobacterium tuberculosis type II dehydroquinase complexed with (1R,4S,5R)-3-(benzo(b)thiophen-5-ylmethoxy)-2-(benzo(b) thiophen-5-ylmethyl)-1,4,5-trihydroxycyclohex-2-enecarboxylate | | Descriptor: | (1R,4S,5R)-3-(BENZO[b]THIOPHEN-5-YL)METHOXY-2-(BENZO[b]THIOPHEN-5-YL)METHYL-1,4,5-TRIHYDROXYCYCLOHEX-2-ENE-1-CARBOXYLATE, 3-DEHYDROQUINATE DEHYDRATASE, SULFATE ION | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Fox, G.C, Tizon, L, Prazeres, V.F.V, Lamb, H, Hawkins, A.R, Ainsa, J.A, Castedo, L, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2011-01-28 | | Release date: | 2011-08-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A prodrug approach for improving antituberculosis activity of potent Mycobacterium tuberculosis type II dehydroquinase inhibitors.
J. Med. Chem., 54, 2011
|
|
2NBU
 
 | |
4ZFC
 
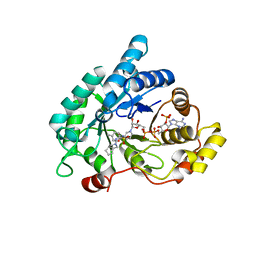 | | Crystal structure of AKR1C3 complexed with glicazide | | Descriptor: | Aldo-keto reductase family 1 member C3, N-[(3aR,6aS)-hexahydrocyclopenta[c]pyrrol-2(1H)-ylcarbamoyl]-4-methylbenzenesulfonamide, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhao, Y, Zhrng, X, Hu, X. | | Deposit date: | 2015-04-21 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | In vitro inhibition of AKR1Cs by sulphonylureas and the structural basis
Chem.Biol.Interact., 240, 2015
|
|
6T98
 
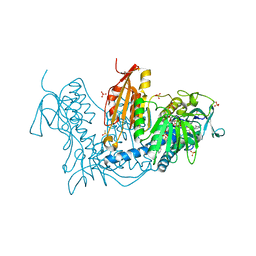 | | Trypanothione Reductase from Leishmania infantum in complex with 9a | | Descriptor: | 4-[3-methyl-1-[4-[4-(2-phenylethyl)-1,3-thiazol-2-yl]-3-(2-piperidin-4-ylethoxy)phenyl]-1,2,3-triazol-3-ium-4-yl]butan-1-amine, DIMETHYL SULFOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Carriles, A.A, Hermoso, J.A. | | Deposit date: | 2019-10-26 | | Release date: | 2020-11-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Identification of 1,2,3-triazolium salt-based inhibitors of Leishmania infantum trypanothione disulfide reductase with enhanced antileishmanial potency in cellulo and increased selectivity.
Eur.J.Med.Chem., 244, 2022
|
|
6T9F
 
 | | CRYSTAL STRUCTURE OF EN ENDOGLUCANASE S308P FROM PENICILLIUM VERRUCULOSUM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoglucanase | | Authors: | Nemashkalov, V, Kravchenko, O, Gabdulkhakov, A, Tischenko, S, Rozhkova, A, Sinitsyn, A. | | Deposit date: | 2019-10-28 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.24847341 Å) | | Cite: | CRYSTAL STRUCTURE OF EN ENDOGLUCANASE S308P FROM PENICILLIUM VERRUCULOSUM
To Be Published
|
|
2NAC
 
 | | HIGH RESOLUTION STRUCTURES OF HOLO AND APO FORMATE DEHYDROGENASE | | Descriptor: | NAD-DEPENDENT FORMATE DEHYDROGENASE, SULFATE ION | | Authors: | Lamzin, V.S, Dauter, Z, Popov, V.O, Harutyunyan, E.H, Wilson, K.S. | | Deposit date: | 1994-07-06 | | Release date: | 1995-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolution structures of holo and apo formate dehydrogenase.
J.Mol.Biol., 236, 1994
|
|
6IP5
 
 | | Cryo-EM structure of the CMV-stalled human 80S ribosome (Structure ii) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Yokoyama, T, Shigematsu, H, Shirouzu, M, Imataka, H, Ito, T. | | Deposit date: | 2018-11-02 | | Release date: | 2019-05-29 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | HCV IRES Captures an Actively Translating 80S Ribosome.
Mol.Cell, 74, 2019
|
|
