3KWC
 
 | | Oxidized, active structure of the beta-carboxysomal gamma-Carbonic Anhydrase, CcmM | | Descriptor: | CHLORIDE ION, Carbon dioxide concentrating mechanism protein, ISOPROPYL ALCOHOL, ... | | Authors: | Kimber, M.S, Castel, S.E, Pena, K.L. | | Deposit date: | 2009-12-01 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the oxidative activation of the carboxysomal {gamma}-carbonic anhydrase, CcmM.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5J6C
 
 | | FMN-dependent Nitroreductase (CDR20291_0767) from Clostridium difficile R20291 | | Descriptor: | FLAVIN MONONUCLEOTIDE, IMIDAZOLE, Putative reductase | | Authors: | Powell, S.M, Wang, B, Hessami, N, Najar, F.Z, Thomas, L.M, West, A.H, Karr, E.A, Richter-Addo, G.B. | | Deposit date: | 2016-04-04 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Crystal structures of two nitroreductases from hypervirulent Clostridium difficile and functionally related interactions with the antibiotic metronidazole.
Nitric Oxide, 60, 2016
|
|
6GM5
 
 | | [FeFe]-hydrogenase HydA1 from Chlamydomonas reinhardtii,variant E141A | | Descriptor: | CHLORIDE ION, Fe-hydrogenase, IRON/SULFUR CLUSTER, ... | | Authors: | Duan, J, Engelbrecht, V, Esselborn, J, Hofmann, E, Winkler, M, Happe, T. | | Deposit date: | 2018-05-24 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystallographic and spectroscopic assignment of the proton transfer pathway in [FeFe]-hydrogenases.
Nat Commun, 9, 2018
|
|
2VVP
 
 | | Crystal structure of Mycobacterium tuberculosis ribose-5-phosphate isomerase B in complex with its substrates ribose 5-phosphate and ribulose 5-phosphate | | Descriptor: | 5-O-phosphono-D-ribose, RIBOSE-5-PHOSPHATE ISOMERASE B, RIBULOSE-5-PHOSPHATE | | Authors: | Kowalinski, E, Roos, A.K, Mariano, S, Salmon, L, Mowbray, S.L. | | Deposit date: | 2008-06-10 | | Release date: | 2008-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | D-Ribose-5-Phosphate Isomerase B from Escherichia Coli is Also a Functional D-Allose-6-Phosphate Isomerase, While the Mycobacterium Tuberculosis Enzyme is not.
J.Mol.Biol., 382, 2008
|
|
5K0Y
 
 | | m48S late-stage initiation complex, purified from rabbit reticulocytes lysates, displaying eIF2 ternary complex and eIF3 i and g subunits relocated to the intersubunit face | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S12, 40S ribosomal protein S21, ... | | Authors: | Simonetti, A, Brito Querido, J, Myasnikov, A.G, Mancera-Martinez, E, Renaud, A, Kuhn, L, Hashem, Y. | | Deposit date: | 2016-05-17 | | Release date: | 2016-07-13 | | Last modified: | 2018-04-18 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | eIF3 Peripheral Subunits Rearrangement after mRNA Binding and Start-Codon Recognition.
Mol.Cell, 63, 2016
|
|
1TTP
 
 | | TRYPTOPHAN SYNTHASE (E.C.4.2.1.20) IN THE PRESENCE OF CESIUM, ROOM TEMPERATURE | | Descriptor: | CESIUM ION, PYRIDOXAL-5'-PHOSPHATE, TRYPTOPHAN SYNTHASE | | Authors: | Rhee, S, Parris, K, Ahmed, S, Miles, E.W, Davies, D.R. | | Deposit date: | 1995-10-11 | | Release date: | 1996-03-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Exchange of K+ or Cs+ for Na+ induces local and long-range changes in the three-dimensional structure of the tryptophan synthase alpha2beta2 complex.
Biochemistry, 35, 1996
|
|
1OE5
 
 | | Xenopus SMUG1, an anti-mutator uracil-DNA Glycosylase | | Descriptor: | 2'-DEOXYURIDINE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-D(*CP*3DRP*GP*GP*AP*CP*TP*3DRP*AP*CP*GP*GP*GP)-3', ... | | Authors: | Wibley, J.E.A, Pearl, L.H. | | Deposit date: | 2003-03-19 | | Release date: | 2003-07-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Specificity of the Vertebrate Anti-Mutator Uracil-DNA Glycosylase Smug1
Mol.Cell, 11, 2003
|
|
5EL6
 
 | | Structure of T. thermophilus 70S ribosome complex with mRNA and tRNALys in the A-site with a U-U mismatch in the first position and antibiotic paromomycin | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Demeshkina, N, Khusainov, I, Yusupov, M, Yusupova, G. | | Deposit date: | 2015-11-04 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Novel base-pairing interactions at the tRNA wobble position crucial for accurate reading of the genetic code.
Nat Commun, 7, 2016
|
|
2AU7
 
 | | The R43Q active site variant of E.coli inorganic pyrophosphatase | | Descriptor: | CHLORIDE ION, Inorganic pyrophosphatase, MANGANESE (II) ION, ... | | Authors: | Samygina, V.R, Avaeva, S.M, Bartunik, H.D. | | Deposit date: | 2005-08-27 | | Release date: | 2006-08-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Reversible inhibition of Escherichia coli inorganic pyrophosphatase by fluoride: trapped catalytic intermediates in cryo-crystallographic studies
J.Mol.Biol., 366, 2007
|
|
5LLP
 
 | | Crystal structure of human carbonic anhydrase isozyme XII with 3-[(1S)-1,2,3,4-Tetrahydronapthalen-1-ylamino)-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide | | Descriptor: | 3-[(1S)-1,2,3,4-Tetrahydronapthalen-1-ylamino)-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, Carbonic anhydrase 12, ZINC ION | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structure correlations with the intrinsic thermodynamics of human carbonic anhydrase inhibitor binding.
PeerJ, 6, 2018
|
|
6M9K
 
 | |
1QG6
 
 | |
5LLO
 
 | | Crystal structure of human carbonic anhydrase isozyme XII with 3-[(1S)-2,3-Dihydro-1H-inden-1-ylamino]-2,5,6-trifluoro-4-[(2-hy-droxyethyl)sulfonyl]benzenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 3-[(1S)-2,3-dihydro-1H-inden-1-ylamino]-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, Carbonic anhydrase 12, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure correlations with the intrinsic thermodynamics of human carbonic anhydrase inhibitor binding.
PeerJ, 6, 2018
|
|
5LLC
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 3-(Methylamino)-2,5,6-trifluoro-4-[(2-phenylethyl)sulfonyl]benzenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 2,3,6-tris(fluoranyl)-5-(methylamino)-4-(2-phenylethylsulfonyl)benzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Manakova, E, Smirnov, A, Grazulis, S. | | Deposit date: | 2016-07-27 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structure correlations with the intrinsic thermodynamics of human carbonic anhydrase inhibitor binding.
PeerJ, 6, 2018
|
|
1U59
 
 | | Crystal Structure of the ZAP-70 Kinase Domain in Complex with Staurosporine | | Descriptor: | STAUROSPORINE, Tyrosine-protein kinase ZAP-70 | | Authors: | Jin, L, Pluskey, S, Petrella, E.C, Cantin, S.M, Gorga, J.C, Rynkiewicz, M.J, Pandey, P, Strickler, J.E, Babine, R.E, Weaver, D.T, Seidl, K.J. | | Deposit date: | 2004-07-27 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Three-dimensional Structure of the ZAP-70 Kinase Domain in Complex with Staurosporine: IMPLICATIONS FOR THE DESIGN OF SELECTIVE INHIBITORS
J.Biol.Chem., 279, 2004
|
|
5LLG
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 4-Propylthiobenzenesulfonamide | | Descriptor: | 4-(propylsulfanyl)benzenesulfonamide, BICINE, Carbonic anhydrase 2, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2016-07-27 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Crystal structure correlations with the intrinsic thermodynamics of human carbonic anhydrase inhibitor binding.
PeerJ, 6, 2018
|
|
6I5M
 
 | | Gamma subunit of the translation initiation factor 2 from Sulfolobus solfataricus in complex with GDP and formate ion | | Descriptor: | FORMIC ACID, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kravchenko, O, Nikonov, O, Gabdulkhakov, A, Stolboushkina, E, Arkhipova, V, Garber, M, Nikonov, S. | | Deposit date: | 2018-11-13 | | Release date: | 2019-01-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The third structural switch in the archaeal translation initiation factor 2 (aIF2) molecule and its possible role in the initiation of GTP hydrolysis and the removal of aIF2 from the ribosome.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
8TJ3
 
 | |
5IRO
 
 | |
2HAK
 
 | | Catalytic and ubiqutin-associated domains of MARK1/PAR-1 | | Descriptor: | Serine/threonine-protein kinase MARK1 | | Authors: | Marx, A, Nugoor, C, Mueller, J, Panneerselvam, S, Mandelkow, E.-M, Mandelkow, E. | | Deposit date: | 2006-06-13 | | Release date: | 2006-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural variations in the catalytic and ubiquitin-associated domains of microtubule-associated protein/microtubule affinity regulating kinase (MARK) 1 and MARK2
J.Biol.Chem., 281, 2006
|
|
5E2X
 
 | | The crystal structure of the C-terminal domain of Ebola (Tai Forest) nucleoprotein | | Descriptor: | NONAETHYLENE GLYCOL, NP | | Authors: | Baker, L.E, Handing, K.B, Derewenda, U, Utepbergenov, D, Derewenda, Z.S. | | Deposit date: | 2015-10-01 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular architecture of the nucleoprotein C-terminal domain from the Ebola and Marburg viruses.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
1KPK
 
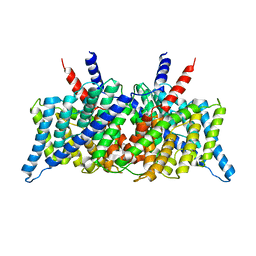 | | Crystal Structure of the ClC Chloride Channel from E. coli | | Descriptor: | putative channel transporter | | Authors: | Dutzler, R, Campbell, E.B, Cadene, M, Chait, B.T, MacKinnon, R. | | Deposit date: | 2001-12-31 | | Release date: | 2002-01-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | X-ray structure of a ClC chloride channel at 3.0 A reveals the molecular basis of
anion selectivity.
Nature, 415, 2002
|
|
1EFR
 
 | | BOVINE MITOCHONDRIAL F1-ATPASE COMPLEXED WITH THE PEPTIDE ANTIBIOTIC EFRAPEPTIN | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BOVINE MITOCHONDRIAL F1-ATPASE SUBUNIT ALPHA, BOVINE MITOCHONDRIAL F1-ATPASE SUBUNIT BETA, ... | | Authors: | Abrahams, J.P, Buchanan, S.K, Van Raaij, M.J, Fearnley, I.M, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 1996-05-24 | | Release date: | 1997-02-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Structure of Bovine F1-ATPase Complexed with the Peptide Antibiotic Efrapeptin.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
5ES3
 
 | | Co-crystal structure of LDH liganded with oxamate | | Descriptor: | L-lactate dehydrogenase A chain, OXAMIC ACID | | Authors: | Nowicki, M.W, Wear, M.A, McNae, I.W, Blackburn, E.A. | | Deposit date: | 2015-11-16 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A Streamlined, Automated Protocol for the Production of Milligram Quantities of Untagged Recombinant Rat Lactate Dehydrogenase A Using AKTAxpressTM.
Plos One, 10, 2015
|
|
1KKA
 
 | |
