8D7U
 
 | |
8D7Y
 
 | |
1L7F
 
 | | Crystal structure of influenza virus neuraminidase in complex with BCX-1812 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(1-ACETYLAMINO-2-ETHYL-BUTYL)-4-GUANIDINO-2-HYDROXY-CYCLOPENTANECARBOXYLIC ACID, ... | | Authors: | Smith, B.J, McKimm-Breshkin, J.L, McDonald, M, Fernley, R.T, Varghese, J.N, Colman, P.M. | | Deposit date: | 2002-03-15 | | Release date: | 2002-05-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural studies of the resistance of influenza virus neuramindase to inhibitors.
J.Med.Chem., 45, 2002
|
|
8D80
 
 | | Cereblon~DDB1 bound to Iberdomide and Ikaros ZF1-2-3 | | Descriptor: | (3S)-3-[4-({4-[(morpholin-4-yl)methyl]phenyl}methoxy)-1-oxo-1,3-dihydro-2H-isoindol-2-yl]piperidine-2,6-dione, DNA damage-binding protein 1, DNA-binding protein Ikaros, ... | | Authors: | Watson, E.R, Lander, G.C. | | Deposit date: | 2022-06-07 | | Release date: | 2022-07-20 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular glue CELMoD compounds are regulators of cereblon conformation.
Science, 378, 2022
|
|
4WPH
 
 | |
6KO9
 
 | | Crystal structure of the Gefitinib Intermediate 1 bound RamR determined with XtaLAB Synergy | | Descriptor: | 4-[(3-chloranyl-4-fluoranyl-phenyl)amino]-7-methoxy-quinazolin-6-ol, Putative regulatory protein, SULFATE ION | | Authors: | Matsumoto, T, Nakashima, R, Yamano, A, Nishino, K. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Development of a structure determination method using a multidrug-resistance regulator protein as a framework.
Biochem.Biophys.Res.Commun., 518, 2019
|
|
6KO8
 
 | | Crystal structure of the Cholic acid bound RamR determined with XtaLAB Synergy | | Descriptor: | CHOLIC ACID, Putative regulatory protein, SULFATE ION | | Authors: | Matsumoto, T, Nakashima, R, Yamano, A, Nishino, K. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Development of a structure determination method using a multidrug-resistance regulator protein as a framework.
Biochem.Biophys.Res.Commun., 518, 2019
|
|
2P88
 
 | | Crystal structure of N-succinyl Arg/Lys racemase from Bacillus cereus ATCC 14579 | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme family protein | | Authors: | Fedorov, A.A, Song, L, Fedorov, E.V, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2007-03-22 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Prediction and assignment of function for a divergent N-succinyl amino acid racemase.
Nat.Chem.Biol., 3, 2007
|
|
6FMQ
 
 | | Keap1 - peptide complex | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ACY-SC1-GLU-THR-GLY-GLU-LEU, ... | | Authors: | Talapatra, S.K, Kozielski, F, Wells, G, Georgakopoulos, N.D. | | Deposit date: | 2018-02-02 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Modified Peptide Inhibitors of the Keap1-Nrf2 Protein-Protein Interaction Incorporating Unnatural Amino Acids.
Chembiochem, 19, 2018
|
|
6TL3
 
 | | Crystal structure of an Estrogen Receptor alpha 8-mer phosphopeptide in complex with 14-3-3sigma stabilized by a Pyrrolidone1 derivative | | Descriptor: | 14-3-3 protein sigma, 5-[(2~{S},3~{R})-3-[(~{R})-azanyl(phenyl)methyl]-2-(4-nitrophenyl)-4,5-bis(oxidanylidene)pyrrolidin-1-yl]-2-oxidanyl-benzoic acid, Estrogen receptor | | Authors: | Andrei, S.A, Bosica, F, Ottmann, C, O'Mahony, G. | | Deposit date: | 2019-11-30 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.455 Å) | | Cite: | Design of Drug-Like Protein-Protein Interaction Stabilizers Guided By Chelation-Controlled Bioactive Conformation Stabilization.
Chemistry, 26, 2020
|
|
4LNL
 
 | | Structure of Escherichia coli Threonine Aldolase in Complex with Allo-Thr | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Low-specificity L-threonine aldolase, ... | | Authors: | Safo, M.K. | | Deposit date: | 2013-07-11 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | On the catalytic mechanism and stereospecificity of Escherichia coli l-threonine aldolase.
Febs J., 281, 2014
|
|
2CBT
 
 | | Crystal structure of the neocarzinostatin 4Tes1 mutant bound testosterone hemisuccinate. | | Descriptor: | NEOCARZINOSTATIN, TESTOSTERONE HEMISUCCINATE | | Authors: | Drevelle, A, Graille, M, Heyd, B, Sorel, I, Ulryck, N, Pecorari, F, Desmadril, M, Van Tilbeurgh, H, Minard, P. | | Deposit date: | 2006-01-06 | | Release date: | 2006-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of in Vitro Evolved Binding Sites on Neocarzinostatin Scaffold Reveal Unanticipated Evolutionary Pathways.
J.Mol.Biol., 358, 2006
|
|
2WHF
 
 | | Interaction of Mycobacterium tuberculosis CYP130 with heterocyclic arylamines | | Descriptor: | 1-(3-METHYLPHENYL)-1H-BENZIMIDAZOL-5-AMINE, PROTOPORPHYRIN IX CONTAINING FE, PUTATIVE CYTOCHROME P450 130 | | Authors: | Podust, L.M, Ouellet, H, von Kries, J.P, Ortiz de Montellano, P.R. | | Deposit date: | 2009-05-04 | | Release date: | 2009-07-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Interaction of Mycobacterium tuberculosis CYP130 with heterocyclic arylamines.
J. Biol. Chem., 284, 2009
|
|
1ADG
 
 | | CRYSTALLOGRAPHIC STUDIES OF TWO ALCOHOL DEHYDROGENASE-BOUND ANALOGS OF THIAZOLE-4-CARBOXAMIDE ADENINE DINUCLEOTIDE (TAD), THE ACTIVE ANABOLITE OF THE ANTITUMOR AGENT TIAZOFURIN | | Descriptor: | ALCOHOL DEHYDROGENASE, BETA-METHYLENE-SELENAZOLE-4-CARBOXYAMIDE-ADENINE DINUCLEOTIDE, ZINC ION | | Authors: | Li, H, Hallows, W.A, Punzi, J.S, Marquez, V.E, Carrell, H.L, Pankiewicz, K.W, Watanabe, K.A, Goldstein, B.M. | | Deposit date: | 1993-10-18 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic studies of two alcohol dehydrogenase-bound analogues of thiazole-4-carboxamide adenine dinucleotide (TAD), the active anabolite of the antitumor agent tiazofurin.
Biochemistry, 33, 1994
|
|
1Z6E
 
 | |
9JC7
 
 | |
6L15
 
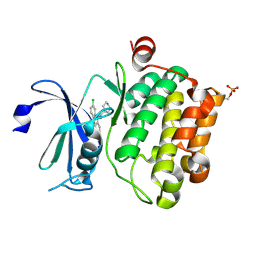 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 7-chloranyl-5-[3-[(3~{S})-piperidin-3-yl]propyl]pyrido[3,4-b][1,4]benzoxazine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
4OHO
 
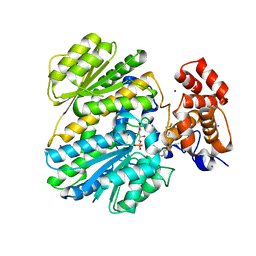 | | Human GKRP bound to AMG-2668 | | Descriptor: | 5-{[(3S)-3-(prop-1-yn-1-yl)-4-{4-[S-(trifluoromethyl)sulfonimidoyl]phenyl}piperazin-1-yl]sulfonyl}pyridin-2-amine, D-SORBITOL-6-PHOSPHATE, GLYCEROL, ... | | Authors: | Jordan, S.R, Chmait, S. | | Deposit date: | 2014-01-17 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Small molecule disruptors of the glucokinase-glucokinase regulatory protein interaction: 3. Structure-activity relationships within the aryl carbinol region of the N-arylsulfonamido-N'-arylpiperazine series.
J.Med.Chem., 57, 2014
|
|
7WML
 
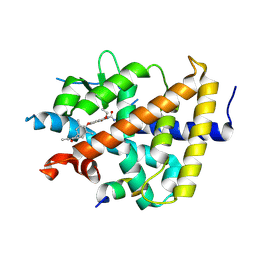 | | A novel chemical derivative(85) of THRB agonist | | Descriptor: | 2-[[7-[4-(3,5-dimethylphenyl)carbonyl-2,6-dimethyl-phenoxy]-1-ethoxy-4-oxidanyl-isoquinolin-3-yl]carbonylamino]ethanoic acid, Isoform Beta-2 of Thyroid hormone receptor beta, Nuclear receptor coactivator 2 | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2022-01-15 | | Release date: | 2022-05-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Discovery of a Highly Selective and H435R-Sensitive Thyroid Hormone Receptor beta Agonist.
J.Med.Chem., 65, 2022
|
|
7WLX
 
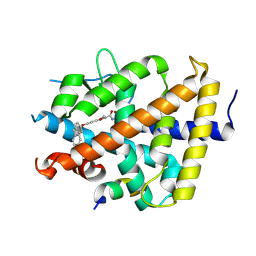 | | A novel chemical derivative(53) of THRB agonist | | Descriptor: | 2-[[1-methoxy-4-oxidanyl-7-[4-(phenylmethyl)phenoxy]isoquinolin-3-yl]carbonylamino]ethanoic acid, Isoform Beta-2 of Thyroid hormone receptor beta, Nuclear receptor coactivator 2 | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Discovery of a Highly Selective and H435R-Sensitive Thyroid Hormone Receptor beta Agonist.
J.Med.Chem., 65, 2022
|
|
1LQ2
 
 | | Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme Exo1 in complex with gluco-phenylimidazole | | Descriptor: | (5R,6R,7S,8S)-5-(HYDROXYMETHYL)-2-PHENYL-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hrmova, M, De Gori, R, Smith, B.J, Vasella, A, Varghese, J.N, Fincher, G.B. | | Deposit date: | 2002-05-09 | | Release date: | 2003-11-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Three-dimensional Structure of the Barley {beta}-D-Glucan Glucohydrolase in Complex with a Transition State Mimic.
J.Biol.Chem., 279, 2004
|
|
5H8N
 
 | | Structure of the human GluN1/GluN2A LBD in complex with NAM | | Descriptor: | 4-[[(4-fluorophenyl)sulfonylamino]methyl]-~{N}-(pyridin-3-ylmethyl)benzamide, CALCIUM ION, GLUTAMIC ACID, ... | | Authors: | Wallweber, H.J.A, Lupardus, P.J. | | Deposit date: | 2015-12-23 | | Release date: | 2016-02-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Positive Allosteric Modulators of GluN2A-Containing NMDARs with Distinct Modes of Action and Impacts on Circuit Function.
Neuron, 89, 2016
|
|
6DHK
 
 | | Bovine glutamate dehydrogenase complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glutamate dehydrogenase 1, mitochondrial | | Authors: | Smith, T.J. | | Deposit date: | 2018-05-20 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural studies on ADP activation of mammalian glutamate dehydrogenase and the evolution of regulation.
Biochemistry, 42, 2003
|
|
6L12
 
 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 4-[(2-chloranylphenoxazin-10-yl)methyl]cyclohexan-1-amine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
1DQK
 
 | | CRYSTAL STRUCTURE OF SUPEROXIDE REDUCTASE IN THE REDUCED STATE AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | FE (II) ION, SUPEROXIDE REDUCTASE | | Authors: | Yeh, A.P, Hu, Y, Jenney Jr, F.E, Adams, M.W.W, Rees, D.C. | | Deposit date: | 2000-01-04 | | Release date: | 2000-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the superoxide reductase from Pyrococcus furiosus in the oxidized and reduced states.
Biochemistry, 39, 2000
|
|
