5D7E
 
 | | Crystal structure of Taf14 YEATS domain in complex with H3K9ac | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, H3K9ac, ... | | Authors: | Andrews, F.H, Shanle, E.K, Strahl, B.D, Kutateladze, T.G. | | Deposit date: | 2015-08-13 | | Release date: | 2015-09-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Association of Taf14 with acetylated histone H3 directs gene transcription and the DNA damage response.
Genes Dev., 29, 2015
|
|
4OFF
 
 | |
4OFG
 
 | | Co-crystal structure of carboxy cGMP binding domain of Plasmodium falciparum PKG with cGMP | | Descriptor: | CGMP-dependent protein kinase, CYCLIC GUANOSINE MONOPHOSPHATE, ETHANOL, ... | | Authors: | Kim, J.J, Sanabria figueroa, E, Kim, C. | | Deposit date: | 2014-01-14 | | Release date: | 2015-01-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of the Carboxyl cGMP Binding Domain of the Plasmodium falciparum cGMP-dependent Protein Kinase Reveal a Novel Capping Triad Crucial for Merozoite Egress.
Plos Pathog., 11, 2015
|
|
5DF9
 
 | | CRYSTAL STRUCTURE OF PENICILLIN-BINDING PROTEIN 3 IN COMPLEX WITH DEACYLATED PRODUCT OF CEFOPERAZONE | | Descriptor: | (2R,5R)-2-[(R)-carboxy{[(2R)-2-{[(4-ethyl-2,3-dioxopiperazin-1-yl)carbonyl]amino}-2-(4-hydroxyphenyl)acetyl]amino}methyl]-5-methyl-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Cell division protein, GLYCEROL, ... | | Authors: | Ren, J, Nettleship, J.E, Males, A, Stuart, D.I, Owens, R.J. | | Deposit date: | 2015-08-26 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of penicillin-binding protein 3 in complexes with azlocillin and cefoperazone in both acylated and deacylated forms.
Febs Lett., 590, 2016
|
|
1B8A
 
 | | ASPARTYL-TRNA SYNTHETASE | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, PROTEIN (ASPARTYL-TRNA SYNTHETASE) | | Authors: | Schmitt, E, Moulinier, L, Thierry, J.-C, Moras, D. | | Deposit date: | 1999-01-27 | | Release date: | 1999-02-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of aspartyl-tRNA synthetase from Pyrococcus kodakaraensis KOD: archaeon specificity and catalytic mechanism of adenylate formation.
EMBO J., 17, 1998
|
|
5DAI
 
 | |
7L6X
 
 | | Crystal structure of the tandem bromodomain (BD1, BD2) of human TAF1 bound to GNE-371 | | Descriptor: | 1,2-ETHANEDIOL, 6-(but-3-en-1-yl)-4-[1-methyl-6-(morpholine-4-carbonyl)-1H-benzimidazol-4-yl]-1,6-dihydro-7H-pyrrolo[2,3-c]pyridin-7-one, Transcription initiation factor TFIID subunit 1 | | Authors: | Karim, M.R, Schonbrunn, E. | | Deposit date: | 2020-12-24 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Discovery of Dual TAF1-ATR Inhibitors and Ligand-Induced Structural Changes of the TAF1 Tandem Bromodomain.
J.Med.Chem., 65, 2022
|
|
4OGW
 
 | | Structure of a human CD38 mutant complexed with NMN | | Descriptor: | ADP-ribosyl cyclase 1, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE | | Authors: | Shewchuk, L.M, Preugschat, F, Carter, L.H, Boros, E.E, Moyer, M.B, Stewart, E.L, Porter, D.J.T. | | Deposit date: | 2014-01-16 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A pre-steady state and steady state kinetic analysis of the N-ribosyl hydrolase activity of hCD157.
Arch.Biochem.Biophys., 564C, 2014
|
|
3NKO
 
 | | Crystal structure of mouse autotaxin in complex with 16:0-LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl hexadecanoate, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nishimasu, H, Ishitani, R, Mihara, E, Takagi, J, Aoki, J, Nureki, O. | | Deposit date: | 2010-06-20 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of autotaxin and insight into GPCR activation by lipid mediators
Nat.Struct.Mol.Biol., 18, 2011
|
|
2ECK
 
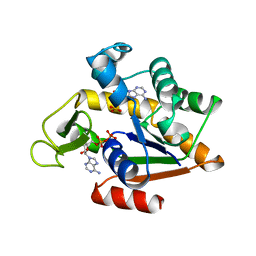 | | STRUCTURE OF PHOSPHOTRANSFERASE | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ADENYLATE KINASE | | Authors: | Berry, M.B, Bilderback, T, Glaser, M, Phillips Jr, G.N. | | Deposit date: | 1996-12-16 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of ADP/AMP complex of Escherichia coli adenylate kinase.
Proteins, 62, 2006
|
|
1JR5
 
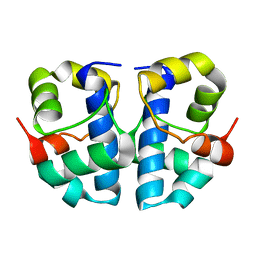 | | Solution Structure of the Anti-Sigma Factor AsiA Homodimer | | Descriptor: | 10 KDA Anti-Sigma Factor | | Authors: | Urbauer, J.L, Simeonov, M.F, Bieber Urbauer, R.J, Adelman, K, Gilmore, J.M, Brody, E.N. | | Deposit date: | 2001-08-10 | | Release date: | 2002-02-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and stability of the anti-sigma factor AsiA: implications for novel functions.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2X55
 
 | | Yersinia Pestis Plasminogen Activator Pla (Native) | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, COAGULASE/FIBRINOLYSIN, SULFATE ION | | Authors: | Eren, E, Murphy, M, Goguen, J, van den Berg, B. | | Deposit date: | 2010-02-05 | | Release date: | 2010-07-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An Active Site Water Network in the Plasminogen Activator Pla from Yersinia Pestis
Structure, 18, 2010
|
|
1ZED
 
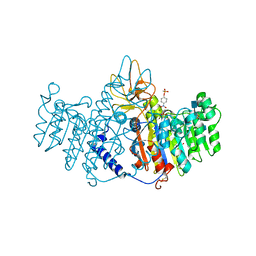 | | Alkaline phosphatase from human placenta in complex with p-nitrophenyl-phosphonate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alkaline phosphatase, CALCIUM ION, ... | | Authors: | Llinas, P, Stura, E.A, Menez, A, Kiss, Z, Stigbrand, T, Millan, J.L, Le Du, M.H. | | Deposit date: | 2005-04-18 | | Release date: | 2005-06-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural Studies of Human Placental Alkaline Phosphatase in Complex with Functional Ligands.
J.Mol.Biol., 350, 2005
|
|
3FEV
 
 | | Crystal structure of the chimeric muscarinic toxin MT7 with loop 1 from MT1. | | Descriptor: | Fusion of Muscarinic toxin 1, Muscarinic m1-toxin1, SULFATE ION | | Authors: | Stura, E.A, Menez, R, Mourier, G, Fruchart-Gaillard, C, Menez, A, Servant, D. | | Deposit date: | 2008-12-01 | | Release date: | 2009-12-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Engineering of three-finger fold toxins creates ligands with original pharmacological profiles for muscarinic and adrenergic receptors.
Plos One, 7, 2012
|
|
2JPA
 
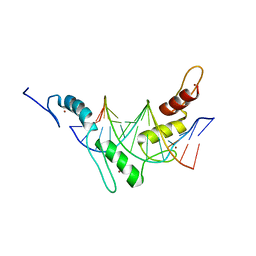 | | Structure of the Wilms Tumor Suppressor Protein Zinc Finger Domain Bound to DNA | | Descriptor: | DNA (5'-D(P*DCP*DAP*DGP*DAP*DCP*DGP*DCP*DCP*DCP*DCP*DCP*DGP*DCP*DG)-3'), DNA (5'-D(P*DCP*DGP*DCP*DGP*DGP*DGP*DGP*DGP*DCP*DGP*DTP*DCP*DTP*DG)-3'), Wilms tumor 1, ... | | Authors: | Stoll, R, Lee, B.M, Debler, E.W, Laity, J.H, Wilson, I.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2007-05-01 | | Release date: | 2007-10-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the wilms tumor suppressor protein zinc finger domain bound to DNA
J.Mol.Biol., 372, 2007
|
|
1T6K
 
 | | Crystal structure of phzF from Pseudomonas fluorescens 2-79 | | Descriptor: | Phenazine biosynthesis protein phzF, SULFATE ION | | Authors: | Parsons, J.F, Song, F, Parsons, L, Calabrese, K, Eisenstein, E, Ladner, J.E. | | Deposit date: | 2004-05-06 | | Release date: | 2004-10-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and function of the phenazine biosynthesis protein PhzF from Pseudomonas fluorescens 2-79
Biochemistry, 43, 2004
|
|
1T7T
 
 | | Crystal structure of the androgen receptor ligand binding domain in complex with 5-alpha dihydrotestosterone | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor | | Authors: | Hur, E, Pfaff, S.J, Payne, E.S, Gron, H, Buehrer, B.M, Fletterick, R.J. | | Deposit date: | 2004-05-10 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Recognition and accommodation at the androgen receptor coactivator binding interface.
Plos Biol., 2, 2004
|
|
3NMA
 
 | | Mutant P169S of Foot-and-mouth disease Virus RNA dependent RNA-polymerase | | Descriptor: | 5'-R(*GP*GP*C)-3', 5'-R(P*CP*C)-3', Genome polyprotein, ... | | Authors: | Agudo, R, Ferrer-Orta, C, Arias, A, Perez-Luque, R, Verdaguer, N, Domingo, E. | | Deposit date: | 2010-06-22 | | Release date: | 2011-05-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A multi-step process of viral adaptation to a mutagenic nucleoside analogue by modulation of transition types leads to extinction-escape.
Plos Pathog., 6, 2010
|
|
5CT5
 
 | | Wild-type Bacillus subtilis lipase A with 10% [BMIM][Cl] | | Descriptor: | 1-butyl-3-methyl-1H-imidazol-3-ium, CHLORIDE ION, SULFATE ION, ... | | Authors: | Nordwald, E.M, Plaks, J.G, Snell, J.R, Sousa, M.C, Kaar, J.L. | | Deposit date: | 2015-07-23 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.747 Å) | | Cite: | Crystallographic Investigation of Imidazolium Ionic Liquid Effects on Enzyme Structure.
Chembiochem, 16, 2015
|
|
1EQ2
 
 | | THE CRYSTAL STRUCTURE OF ADP-L-GLYCERO-D-MANNOHEPTOSE 6-EPIMERASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE-GLUCOSE, ADP-L-GLYCERO-D-MANNOHEPTOSE 6-EPIMERASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Deacon, A.M, Ni, Y.S, Coleman Jr, W.G, Ealick, S.E. | | Deposit date: | 2000-03-31 | | Release date: | 2000-11-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of ADP-L-glycero-D-mannoheptose 6-epimerase: catalysis with a twist.
Structure Fold.Des., 8, 2000
|
|
1NVD
 
 | | Crystal structure of 3-dehydroquinate synthase (DHQS) in complex with ZN2+ and carbaphosphonate | | Descriptor: | 3-DEHYDROQUINATE SYNTHASE, CHLORIDE ION, ZINC ION, ... | | Authors: | Nichols, C.E, Ren, J, Lamb, H.K, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2003-02-03 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Ligand-induced Conformational Changes and a Mechanism for Domain Closure in Aspergillus nidulans Dehydroquinate Synthase
J.MOL.BIOL., 327, 2003
|
|
2WGB
 
 | | Crystal structure of the TetR-like transcriptional regulator LfrR from Mycobacterium smegmatis | | Descriptor: | SULFATE ION, TETR FAMILY TRANSCRIPTIONAL REPRESSOR LFRR | | Authors: | Bellinzoni, M, Buroni, S, Riccardi, G, De Rossi, E, Alzari, P.M. | | Deposit date: | 2009-04-16 | | Release date: | 2009-10-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Plasticity and Distinct Drug-Binding Modes of Lfrr, a Mycobacterial Efflux Pump Regulator.
J.Bacteriol., 191, 2009
|
|
7L9K
 
 | | Crystal structure of the second bromodomain (BD2) of human BRD2 bound to LRRK2-IN-1 | | Descriptor: | 1,4-BUTANEDIOL, 2-[(2-methoxy-4-{[4-(4-methylpiperazin-1-yl)piperidin-1-yl]carbonyl}phenyl)amino]-5,11-dimethyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain-containing protein 2 | | Authors: | Karim, M.R, Bikowitz, M.J, Schonbrunn, E. | | Deposit date: | 2021-01-04 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7LB4
 
 | |
2JVG
 
 | | Structure of C3-binding domain 4 of Staphylococcus aureus protein Sbi | | Descriptor: | IgG-binding protein SBI | | Authors: | Upadhyay, A, Burman, J, Clark, E.A, van den Elsen, J.M.H, Bagby, S. | | Deposit date: | 2007-09-20 | | Release date: | 2008-06-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of the C3 binding region of Staphylococcus aureus immune subversion protein Sbi.
J.Biol.Chem., 283, 2008
|
|
