9EYJ
 
 | |
9C8V
 
 | | Human DNA polymerase alpha/primase - CHAPSO (4 mM) | | Descriptor: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase large subunit, ... | | Authors: | Abe, K.M, Li, G, He, Q, Grant, T, Lim, C. | | Deposit date: | 2024-06-13 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Human DNA polymerase alpha/primase - CHAPSO (4 mM)
To Be Published
|
|
7A83
 
 | |
9C7V
 
 | | Structure of the human BOS:human EMC complex in GDN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BOS complex subunit NOMO2, ... | | Authors: | Nguyen, V.N, Tomaleri, G.P, Voorhees, R.M. | | Deposit date: | 2024-06-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Role of a holo-insertase complex in the biogenesis of biophysically diverse ER membrane proteins.
Mol.Cell, 84, 2024
|
|
8DOL
 
 | | Mechanism of regulation of the Helicobacter pylori Cagbeta ATPase by CagZ | | Descriptor: | Cag pathogenicity island protein (Cag5), DI(HYDROXYETHYL)ETHER, SULFATE ION | | Authors: | Wu, X, Zhao, Y, Yang, W, Sun, L, Ye, X, Jiang, M, Wang, Q, Wang, Q, Zhang, X, Wu, Y. | | Deposit date: | 2022-07-13 | | Release date: | 2023-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism of regulation of the Helicobacter pylori Cag beta ATPase by CagZ.
Nat Commun, 14, 2023
|
|
9CYP
 
 | | Crystal structure of I19V mutant human PTP1B (PTPN1) at room temperature (298 K) | | Descriptor: | BETA-MERCAPTOETHANOL, MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Ebrahim, A, Perdikari, A, Woods, V.A, Lawler, K, Bounds, R, Singh, N.I, Mehlman, T, Riley, B.T, Sharma, S, Morris, J.W, Keogh, J.M, Henning, E, Smith, M, Farooqi, I.S, Keedy, D.A. | | Deposit date: | 2024-08-02 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structures of human PTP1B variants reveal allosteric sites to target for weight loss therapy.
Biorxiv, 2024
|
|
6LQW
 
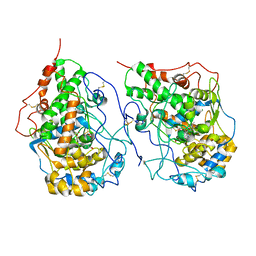 | | Crystal structure of a dimeric yak lactoperoxidase at 2.59 A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Viswanathan, V, Pandey, S.N, Ahmad, N, Rani, C, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2020-01-14 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a dimeric yak lactoperoxidase at 2.59 A resolution.
To Be Published
|
|
6IAL
 
 | | Porcine E.coli heat-labile enterotoxin B-pentamer in complex with Lacto-N-neohexaose | | Descriptor: | CALCIUM ION, GLYCEROL, Heat-labile enterotoxin B chain, ... | | Authors: | Heim, J.B, Heggelund, J.E, Krengel, U. | | Deposit date: | 2018-11-27 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Specificity ofEscherichia coliHeat-Labile Enterotoxin Investigated by Single-Site Mutagenesis and Crystallography.
Int J Mol Sci, 20, 2019
|
|
9F5H
 
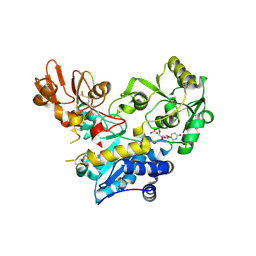 | | Crystal structure of MGAT5 bump-and-hole mutant in complex with UDP and M592 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Secreted alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase A, ... | | Authors: | Liu, Y, Bineva-Todd, G, Meek, R, Mazo, L, Piniello, B, Moroz, O.V, Begum, N, Roustan, C, Tomita, S, Kjaer, S, Rovira, C, Davies, G.J, Schumann, B. | | Deposit date: | 2024-04-28 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A Bioorthogonal Precision Tool for Human N -Acetylglucosaminyltransferase V.
J.Am.Chem.Soc., 146, 2024
|
|
6P3R
 
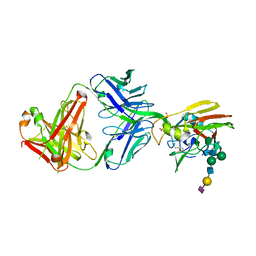 | |
9CVE
 
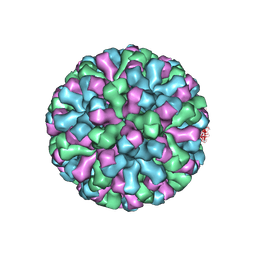 | |
7M1Q
 
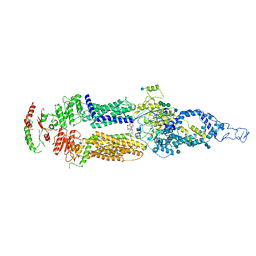 | | Human ABCA4 structure in complex with N-ret-PE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Retinal-specific phospholipid-transporting ATPase ABCA4, ... | | Authors: | Scortecci, J.F, Van Petegem, F, Molday, R.S. | | Deposit date: | 2021-03-14 | | Release date: | 2021-09-08 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Cryo-EM structures of the ABCA4 importer reveal mechanisms underlying substrate binding and Stargardt disease.
Nat Commun, 12, 2021
|
|
9C9Y
 
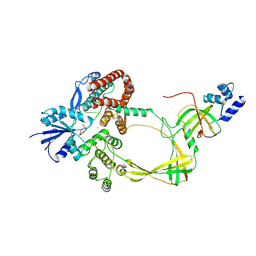 | | Human TOP3B-TDRD3 core complex in DNA pre-cleavage state | | Descriptor: | DNA (5'-D(P*AP*CP*TP*AP*AP*AP*AP*T)-3'), DNA topoisomerase 3-beta-1, MANGANESE (II) ION, ... | | Authors: | Yang, X, Chen, X, Yang, W, Pommier, Y. | | Deposit date: | 2024-06-16 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural Insights into human Topoisomerase 3-beta DNA and RNA catalytic cycles and topo-gate dynamics
To Be Published
|
|
6IMW
 
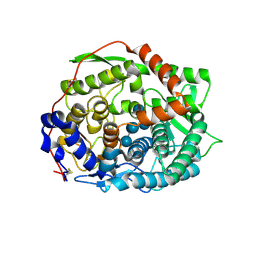 | | The complex structure of endo-beta-1,2-glucanase mutant (E262Q) from Talaromyces funiculosus with beta-1,2-glucan | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, Endo-beta-1,2-glucanase, ... | | Authors: | Tanaka, N, Nakajima, M, Narukawa-Nara, M, Matsunaga, H, Kamisuki, S, Aramasa, H, Takahashi, Y, Sugimoto, N, Abe, K, Miyanaga, A, Yamashita, T, Sugawara, F, Kamakura, T, Komba, S, Nakai, H, Taguchi, H. | | Deposit date: | 2018-10-23 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification, characterization, and structural analyses of a fungal endo-beta-1,2-glucanase reveal a new glycoside hydrolase family.
J.Biol.Chem., 294, 2019
|
|
9BXI
 
 | |
9CIP
 
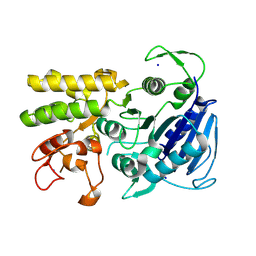 | | MicroED structure of the C11 cysteine protease clostripain | | Descriptor: | Clostripain, SODIUM ION | | Authors: | Ruma, Y.N, Bu, G, Hattne, J, Gonen, T. | | Deposit date: | 2024-07-03 | | Release date: | 2024-08-14 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.5 Å) | | Cite: | MicroED structure of the C11 cysteine protease clostripain.
J Struct Biol X, 10, 2024
|
|
9CYM
 
 | | Structure of LAG3 bound to the MHC class II molecule I-A(b) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Class-II-associated invariant chain peptide, H-2 class II histocompatibility antigen, ... | | Authors: | Ming, Q, Antfolk, D, Tran, T.H, Luca, V.C. | | Deposit date: | 2024-08-02 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.84 Å) | | Cite: | Structural basis for mouse LAG3 interactions with the MHC class II molecule I-A b.
Nat Commun, 15, 2024
|
|
9FDV
 
 | | Crystal Structure of reduced NuoEF variant R66G(NuoF) from Aquifex aeolicus | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ... | | Authors: | Wohlwend, D, Friedrich, T, Goeppert-Asadollahpour, S. | | Deposit date: | 2024-05-17 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.986 Å) | | Cite: | Structural robustness of the NADH binding site in NADH:ubiquinone oxidoreductase (complex I).
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
6M77
 
 | |
8E80
 
 | | Structure of LRRK2-CHK1 10-pt. mutant complex with heteroaryl-1H-indazole LRRK2 inhibitor 14 | | Descriptor: | 2-[(1r,4r)-2-{(6P)-6-[(6M)-6-(1H-pyrazol-5-yl)-1H-indazol-1-yl]pyrimidin-4-yl}-2-azabicyclo[2.1.1]hexan-4-yl]propan-2-ol, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L. | | Deposit date: | 2022-08-25 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Discovery and Optimization of Potent, Selective, and Brain-Penetrant 1-Heteroaryl-1 H -Indazole LRRK2 Kinase Inhibitors for the Treatment of Parkinson's Disease.
J.Med.Chem., 65, 2022
|
|
3C6Q
 
 | |
9CIJ
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23E/L36R at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Zhang, Y, Schlessman, L.J, Siegler, M.A, Garcia-Moreno E, B. | | Deposit date: | 2024-07-03 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23E/L36R at cryogenic temperature
To Be Published
|
|
5JFD
 
 | | Thrombin in complex with (S)-N-(2-(aminomethyl)-5-chlorobenzyl)-1-((benzylsulfonyl)-D-arginyl)pyrrolidine-2-carboxamide | | Descriptor: | (2S)-N-[[2-(aminomethyl)-5-chloro-phenyl]methyl]-1-[(2R)-5-carbamimidamido-2-(phenylmethylsulfonylamino)pentanoyl]pyrrolidine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Sandner, A, Heine, A, Klebe, G. | | Deposit date: | 2016-04-19 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Strategies for Late-Stage Optimization: Profiling Thermodynamics by Preorganization and Salt Bridge Shielding.
J.Med.Chem., 62, 2019
|
|
8E81
 
 | | Structure of LRRK2-CHK1 10-pt. mutant complex with heteroaryl-1H-indazole LRRK2 inhibitor 25 | | Descriptor: | (1S)-1-[(1P)-1-{6-[(3R)-3-(2-hydroxypropan-2-yl)pyrrolidin-1-yl]pyrimidin-4-yl}-1H-indazol-6-yl]spiro[2.2]pentane-1-carbonitrile, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L. | | Deposit date: | 2022-08-25 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery and Optimization of Potent, Selective, and Brain-Penetrant 1-Heteroaryl-1 H -Indazole LRRK2 Kinase Inhibitors for the Treatment of Parkinson's Disease.
J.Med.Chem., 65, 2022
|
|
9EM5
 
 | | OPR3 variant Y364P in its monomeric form | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 12-oxophytodienoate reductase 3, CHLORIDE ION, ... | | Authors: | Bijelic, A, Macheroux, P, Kerschbaumer, B. | | Deposit date: | 2024-03-07 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Analysis of homodimer formation in 12-oxophytodienoate reductase 3 in solutio and crystallo challenges the physiological role of the dimer.
Sci Rep, 14, 2024
|
|
