3ZZH
 
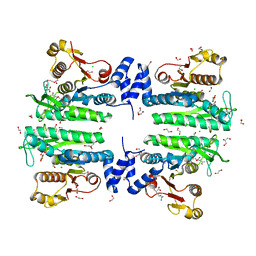 | | Crystal structure of the amino acid kinase domain from Saccharomyces cerevisiae acetylglutamate kinase in complex with its feed- back inhibitor L-arginine | | Descriptor: | 1,2-ETHANEDIOL, ACETYLGLUTAMATE KINASE, ARGININE, ... | | Authors: | de Cima, S, Gil-Ortiz, F, Crabeel, M, Fita, I, Rubio, V. | | Deposit date: | 2011-09-01 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insight on an Arginine Synthesis Metabolon from the Tetrameric Structure of Yeast Acetylglutamate Kinase
Plos One, 7, 2012
|
|
6JQO
 
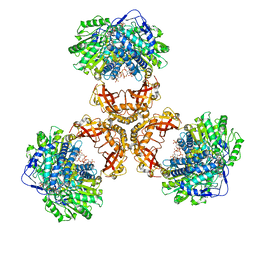 | | Structure of PaaZ, a bifunctional enzyme in complex with NADP+ and CCoA | | Descriptor: | Bifunctional protein PaaZ, CROTONYL COENZYME A, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Gakher, L, Vinothkumar, K.R, Katagihallimath, N, Sowdhamini, R, Sathyanarayanan, N, Cannone, G. | | Deposit date: | 2019-03-31 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis for metabolite channeling in a ring opening enzyme of the phenylacetate degradation pathway.
Nat Commun, 10, 2019
|
|
1GWO
 
 | |
7DLR
 
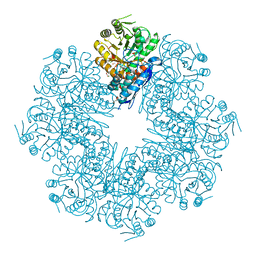 | | Mycobacterium tuberculosis enolase mutant - E163A | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Ahmad, M, Biswal, B.K. | | Deposit date: | 2020-11-30 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural snapshots of Mycobacterium tuberculosis enolase reveal dual mode of 2PG binding and its implication in enzyme catalysis.
Iucrj, 10, 2023
|
|
7UOM
 
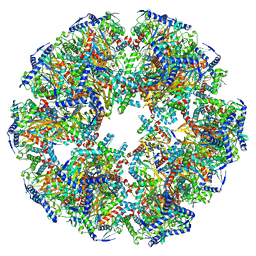 | | Endogenous dihydrolipoamide acetyltransferase (E2) core of pyruvate dehydrogenase complex from bovine kidney | | Descriptor: | Acetyltransferase component of pyruvate dehydrogenase complex | | Authors: | Liu, S, Xia, X, Zhen, J, Li, Z.H, Zhou, Z.H. | | Deposit date: | 2022-04-13 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures and comparison of endogenous 2-oxoglutarate and pyruvate dehydrogenase complexes from bovine kidney.
Cell Discov, 8, 2022
|
|
7CLK
 
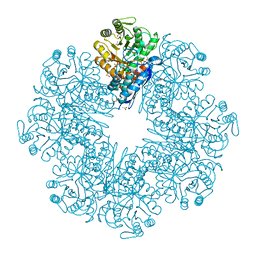 | | Mycobacterium tuberculosis enolase in complex with alternate 2-phosphoglycerate | | Descriptor: | 1,2-ETHANEDIOL, 2-PHOSPHOGLYCERIC ACID, ACETATE ION, ... | | Authors: | Ahmad, M, Jha, B, Tiwari, S, Pal, R.K, Biswal, B.K. | | Deposit date: | 2020-07-21 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural snapshots of Mycobacterium tuberculosis enolase reveal dual mode of 2PG binding and its implication in enzyme catalysis.
Iucrj, 10, 2023
|
|
5I59
 
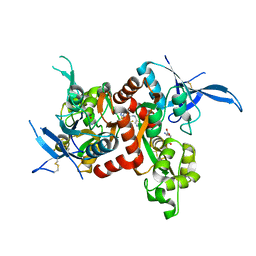 | | Glutamate- and glycine-bound GluN1/GluN2A agonist binding domains with MPX 007 | | Descriptor: | 5-({[(3,4-difluorophenyl)sulfonyl]amino}methyl)-6-methyl-N-[(2-methyl-4H-1lambda~4~,3-thiazol-5-yl)methyl]pyrazine-2-carboxamide, GLUTAMIC ACID, GLYCINE, ... | | Authors: | Mou, T.-C, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2016-02-14 | | Release date: | 2016-09-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Basis for Negative Allosteric Modulation of GluN2A-Containing NMDA Receptors.
Neuron, 91, 2016
|
|
1A7T
 
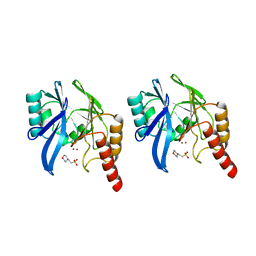 | | METALLO-BETA-LACTAMASE WITH MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, METALLO-BETA-LACTAMASE, SODIUM ION, ... | | Authors: | Fitzgerald, P.M.D, Wu, J.K, Toney, J.H. | | Deposit date: | 1998-03-17 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Unanticipated inhibition of the metallo-beta-lactamase from Bacteroides fragilis by 4-morpholineethanesulfonic acid (MES): a crystallographic study at 1.85-A resolution.
Biochemistry, 37, 1998
|
|
5OCU
 
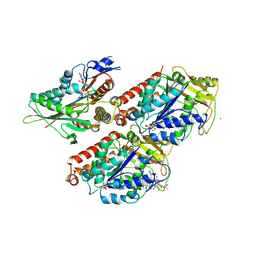 | | Molecular basis of human kinesin-8 function and inhibition | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein KIF18A, ... | | Authors: | Locke, J, Joseph, A.P, Topf, M, Moores, C.A. | | Deposit date: | 2017-07-03 | | Release date: | 2017-10-25 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Structural basis of human kinesin-8 function and inhibition.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6CIG
 
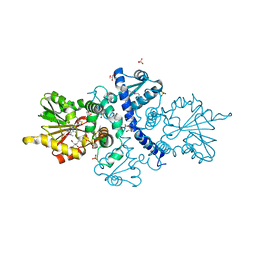 | | CRYSTAL STRUCTURE ANALYSIS OF SELENOMETHIONINE SUBSTITUTED ISOFLAVONE O-METHYLTRANSFERASE | | Descriptor: | GLYCEROL, Isoflavone-7-O-methyltransferase 8, N-(TRIS(HYDROXYMETHYL)METHYL)-3-AMINOPROPANESULFONIC ACID, ... | | Authors: | Zubieta, C, Dixon, R.A, Shabalin, I.G, Kowiel, M, Porebski, P.J, Noel, J.P. | | Deposit date: | 2018-02-23 | | Release date: | 2018-03-07 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of two natural product methyltransferases reveal the basis for substrate specificity in plant O-methyltransferases.
Nat. Struct. Biol., 8, 2001
|
|
1WXD
 
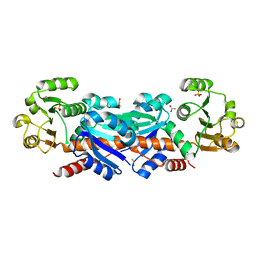 | |
5WTA
 
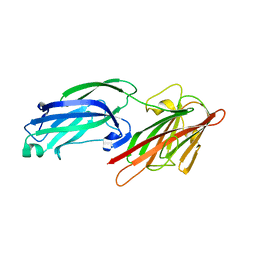 | | Crystal Structure of Staphylococcus aureus SdrE apo form | | Descriptor: | Serine-aspartate repeat-containing protein E | | Authors: | Wu, M, Zhang, Y, Hang, T, Wang, C, Yang, Y, Zang, J, Zhang, M, Zhang, X. | | Deposit date: | 2016-12-10 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Staphylococcus aureus SdrE captures complement factor H's C-terminus via a novel 'close, dock, lock and latch' mechanism for complement evasion
Biochem. J., 474, 2017
|
|
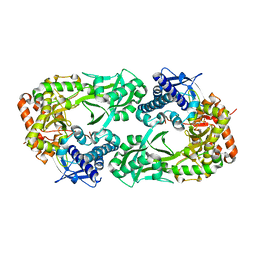
6CN7
 
 | |
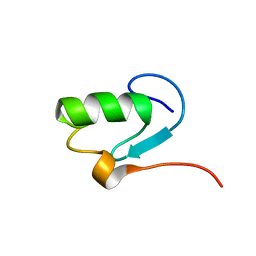
1N87
 
 | |
5I56
 
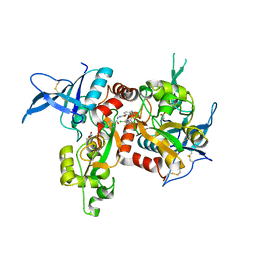 | | Agonist-bound GluN1/GluN2A agonist binding domains with TCN201 | | Descriptor: | GLUTAMIC ACID, GLYCINE, Glutamate receptor ionotropic, ... | | Authors: | Mou, T.-C, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2016-02-14 | | Release date: | 2016-09-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural Basis for Negative Allosteric Modulation of GluN2A-Containing NMDA Receptors.
Neuron, 91, 2016
|
|
5LOV
 
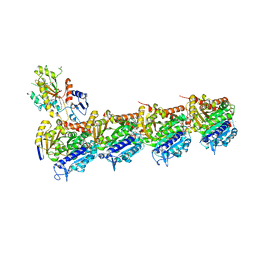 | | DZ-2384 tubulin complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, DZ 2384, ... | | Authors: | Prota, A.E, Steinmetz, M.O, Shore, G.C, Brouhard, G, Roulston, A. | | Deposit date: | 2016-08-10 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The synthetic diazonamide DZ-2384 has distinct effects on microtubule curvature and dynamics without neurotoxicity.
Sci Transl Med, 8, 2016
|
|
5FIT
 
 | | FHIT-SUBSTRATE ANALOG | | Descriptor: | FRAGILE HISTIDINE TRIAD PROTEIN, PHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER | | Authors: | Lima, C.D, Klein, M.G, Hendrickson, W.A. | | Deposit date: | 1997-09-25 | | Release date: | 1998-03-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based analysis of catalysis and substrate definition in the HIT protein family.
Science, 278, 1997
|
|
2VWP
 
 | | Haloferax mediterranei glucose dehydrogenase in complex with NADPH and Zn. | | Descriptor: | GLUCOSE DEHYDROGENASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ZINC ION | | Authors: | Baker, P.J, Britton, K.L, Fisher, M, Esclapez, J, Pire, C, Bonete, M.J, Ferrer, J, Rice, D.W. | | Deposit date: | 2008-06-26 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Active site dynamics in the zinc-dependent medium chain alcohol dehydrogenase superfamily.
Proc. Natl. Acad. Sci. U.S.A., 106, 2009
|
|
7UJX
 
 | | Structure of cAMP-dependent protein kinase using a MD-MX procedure, produced using 2.4 Angstrom data | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Wych, D.C, Aoto, P.C, Wall, M.E. | | Deposit date: | 2022-03-31 | | Release date: | 2022-12-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular-dynamics simulation methods for macromolecular crystallography.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
5I58
 
 | | GLUTAMATE- AND GLYCINE-BOUND GLUN1/GLUN2A AGONIST BINDING DOMAINS WITH MPX-004 | | Descriptor: | 5-({[(3-chloro-4-fluorophenyl)sulfonyl]amino}methyl)-N-[(2-methyl-1,3-thiazol-5-yl)methyl]pyrazine-2-carboxamide, GLUTAMIC ACID, GLYCINE, ... | | Authors: | Mou, T.-C, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2016-02-14 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural Basis for Negative Allosteric Modulation of GluN2A-Containing NMDA Receptors.
Neuron, 91, 2016
|
|
5IA0
 
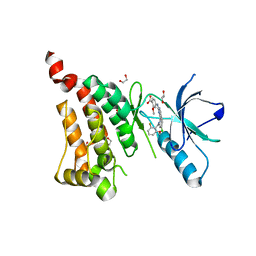 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with alisertib (MLN8237) | | Descriptor: | 1,2-ETHANEDIOL, Ephrin type-A receptor 2, alisertib | | Authors: | Kudlinzki, D, Linhard, V.L, Gande, S.L, Sreeramulu, S, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2016-02-21 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Chemical Proteomics and Structural Biology Define EPHA2 Inhibition by Clinical Kinase Drugs.
ACS Chem. Biol., 11, 2016
|
|
7ZM0
 
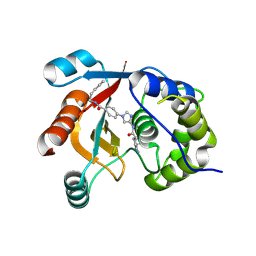 | | Structure of UCHL1 in complex with GK13S inhibitor | | Descriptor: | (3S)-1-(iminomethyl)-N-[1-[4-(pent-4-ynylcarbamoyl)phenyl]imidazol-4-yl]pyrrolidine-3-carboxamide, Ubiquitin carboxyl-terminal hydrolase isozyme L1 | | Authors: | Grethe, C, Gersch, M. | | Deposit date: | 2022-04-18 | | Release date: | 2022-09-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural basis for specific inhibition of the deubiquitinase UCHL1.
Nat Commun, 13, 2022
|
|
5I57
 
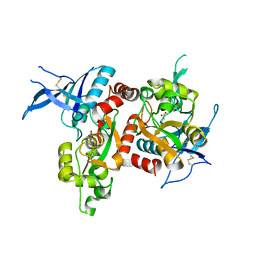 | | Glutamate- and glycine-bound GluN1/GluN2A agonist binding domains | | Descriptor: | GLUTAMIC ACID, GLYCINE, Glutamate receptor ionotropic, ... | | Authors: | Mou, T.-C, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2016-02-14 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Negative Allosteric Modulation of GluN2A-Containing NMDA Receptors.
Neuron, 91, 2016
|
|
2O68
 
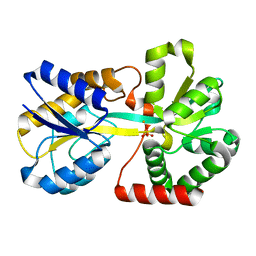 | | Crystal Structure of Haemophilus influenzae Q58L mutant FbpA | | Descriptor: | FE (III) ION, Iron-utilization periplasmic protein, PHOSPHATE ION | | Authors: | Shouldice, S.R, Tari, L.W. | | Deposit date: | 2006-12-07 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The role of the synergistic phosphate anion in iron transport by the periplasmic iron-binding protein from Haemophilus influenzae.
Biochem.J., 403, 2007
|
|
7V0G
 
 | | Structure of cAMP-dependent protein kinase using a MD-MX procedure, produced using 1.63 Angstrom data | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Wych, D.C, Aoto, P.C, Wall, M.E. | | Deposit date: | 2022-05-10 | | Release date: | 2022-12-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Molecular-dynamics simulation methods for macromolecular crystallography.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
