1BHT
 
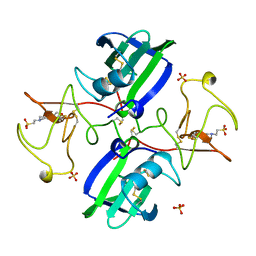 | | NK1 FRAGMENT OF HUMAN HEPATOCYTE GROWTH FACTOR | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEPATOCYTE GROWTH FACTOR, SULFATE ION | | Authors: | Ultsch, M.H, Lokker, N.A, Godowski, P.J, De Vos, A.M. | | Deposit date: | 1998-06-10 | | Release date: | 1998-11-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the NK1 fragment of human hepatocyte growth factor at 2.0 A resolution.
Structure, 6, 1998
|
|
1B49
 
 | | DCMP HYDROXYMETHYLASE FROM T4 (PHOSPHATE-BOUND) | | Descriptor: | PHOSPHATE ION, PROTEIN (DEOXYCYTIDYLATE HYDROXYMETHYLASE) | | Authors: | Song, H.K, Sohn, S.H, Suh, S.W. | | Deposit date: | 1999-01-06 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of deoxycytidylate hydroxymethylase from bacteriophage T4, a component of the deoxyribonucleoside triphosphate-synthesizing complex.
EMBO J., 18, 1999
|
|
1B7B
 
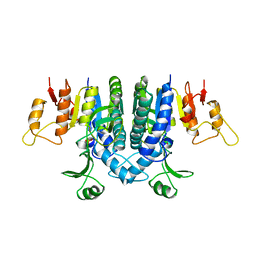 | | Carbamate kinase from Enterococcus faecalis | | Descriptor: | CARBAMATE KINASE, SULFATE ION | | Authors: | Marina, A, Alzari, P.M, Bravo, J, Uriarte, M, Barcelona, B, Fita, I, Rubio, V. | | Deposit date: | 1999-01-20 | | Release date: | 2000-01-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Carbamate kinase: New structural machinery for making carbamoyl phosphate, the common precursor of pyrimidines and arginine.
Protein Sci., 8, 1999
|
|
1BIT
 
 | |
1BGE
 
 | |
1B8D
 
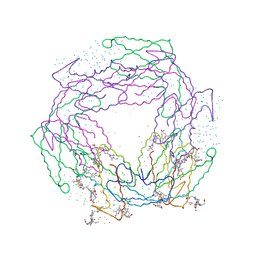 | | CRYSTAL STRUCTURE OF A PHYCOUROBILIN-CONTAINING PHYCOERYTHRIN | | Descriptor: | PHYCOERYTHROBILIN, PHYCOUROBILIN, PROTEIN (RHODOPHYTAN PHYCOERYTHRIN (ALPHA CHAIN)), ... | | Authors: | Ritter, S, Hiller, R.G, Wrench, P.M, Welte, W, Diederichs, K. | | Deposit date: | 1999-01-29 | | Release date: | 1999-02-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a phycourobilin-containing phycoerythrin at 1.90-A resolution.
J.Struct.Biol., 126, 1999
|
|
1BE6
 
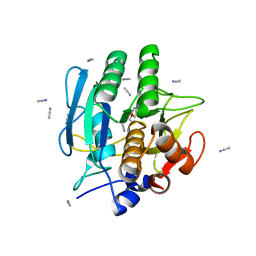 | | TRANS-CINNAMOYL-SUBTILISIN IN ANHYDROUS ACETONITRILE | | Descriptor: | ACETONITRILE, CALCIUM ION, PHENYLETHYLENECARBOXYLIC ACID, ... | | Authors: | Schmitke, J.L, Stern, L.J, Klibanov, A.M. | | Deposit date: | 1998-05-20 | | Release date: | 1998-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Comparison of x-ray crystal structures of an acyl-enzyme intermediate of subtilisin Carlsberg formed in anhydrous acetonitrile and in water.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1BF3
 
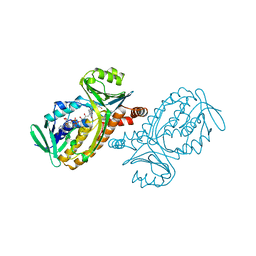 | | P-HYDROXYBENZOATE HYDROXYLASE (PHBH) MUTANT WITH CYS 116 REPLACED BY SER (C116S) AND ARG 42 REPLACED BY LYS (R42K), IN COMPLEX WITH FAD AND 4-HYDROXYBENZOIC ACID | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE, P-HYDROXYBENZOIC ACID | | Authors: | Eppink, M.H.M, Schreuder, H.A, Van Berkel, W.J.H. | | Deposit date: | 1998-05-26 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Lys42 and Ser42 variants of p-hydroxybenzoate hydroxylase from Pseudomonas fluorescens reveal that Arg42 is essential for NADPH binding.
Eur.J.Biochem., 253, 1998
|
|
1B9Z
 
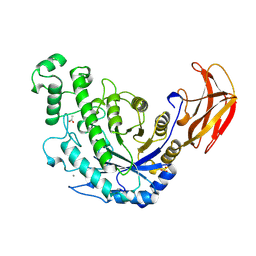 | | BACILLUS CEREUS BETA-AMYLASE COMPLEXED WITH MALTOSE | | Descriptor: | ACETATE ION, CALCIUM ION, PROTEIN (BETA-AMYLASE), ... | | Authors: | Mikami, B, Adachi, M, Kage, T, Sarikaya, E, Nanmori, T, Shinke, R, Utsumi, S. | | Deposit date: | 1999-03-06 | | Release date: | 1999-03-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of raw starch-digesting Bacillus cereus beta-amylase complexed with maltose.
Biochemistry, 38, 1999
|
|
1BB5
 
 | |
1LE5
 
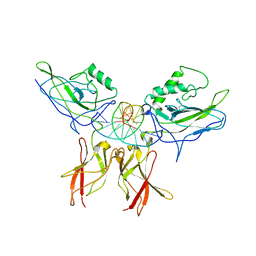 | | Crystal structure of a NF-kB heterodimer bound to an IFNb-kB | | Descriptor: | 5'-D(*AP*AP*GP*GP*AP*AP*TP*TP*TP*CP*CP*C)-3', 5'-D(*TP*GP*GP*GP*AP*AP*AP*TP*TP*CP*CP*T)-3', Nuclear factor NF-kappa-B p50 subunit, ... | | Authors: | Berkowitz, B, Huang, D.B, Chen-Park, F.E, Sigler, P.B, Ghosh, G. | | Deposit date: | 2002-04-09 | | Release date: | 2003-04-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The X-ray crystal structure of the
NF-kB p50/p65 heterodimer bound
to the Interferon beta-kB site
J.Biol.Chem., 277, 2002
|
|
1BBU
 
 | | LYSYL-TRNA SYNTHETASE (LYSS) COMPLEXED WITH LYSINE | | Descriptor: | LYSINE, PROTEIN (LYSYL-TRNA SYNTHETASE) | | Authors: | Onesti, S, Desogus, G, Brevet, A, Chen, J, Plateau, P, Blanquet, S, Brick, P. | | Deposit date: | 1998-04-24 | | Release date: | 2000-11-10 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural studies of lysyl-tRNA synthetase: conformational changes induced by substrate binding.
Biochemistry, 39, 2000
|
|
1BGS
 
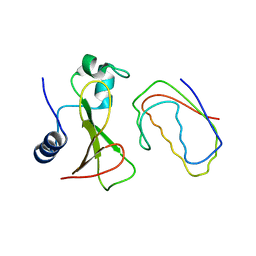 | | RECOGNITION BETWEEN A BACTERIAL RIBONUCLEASE, BARNASE, AND ITS NATURAL INHIBITOR, BARSTAR | | Descriptor: | BARNASE, BARSTAR | | Authors: | Guillet, V, Lapthorn, A, Mauguen, Y. | | Deposit date: | 1993-11-02 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Recognition between a bacterial ribonuclease, barnase, and its natural inhibitor, barstar.
Structure, 1, 1993
|
|
1BG1
 
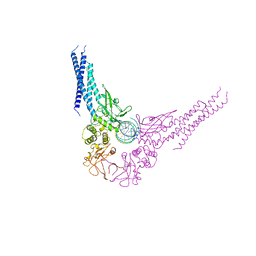 | | TRANSCRIPTION FACTOR STAT3B/DNA COMPLEX | | Descriptor: | DNA (5'-D(*TP*GP*CP*AP*TP*TP*TP*CP*CP*CP*GP*TP*AP*AP*AP*TP*CP*T)-3'), PROTEIN (TRANSCRIPTION FACTOR STAT3B) | | Authors: | Becker, S, Groner, B, Muller, C.W. | | Deposit date: | 1998-06-03 | | Release date: | 1999-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Three-dimensional structure of the Stat3beta homodimer bound to DNA.
Nature, 394, 1998
|
|
1BIF
 
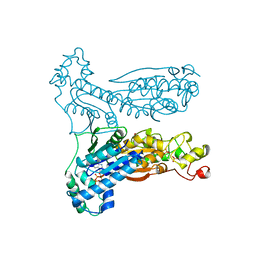 | | 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE BIFUNCTIONAL ENZYME COMPLEXED WITH ATP-G-S AND PHOSPHATE | | Descriptor: | 6-PHOSPHOFRUCTO-2-KINASE/ FRUCTOSE-2,6-BISPHOSPHATASE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Hasemann, C.A, Deisenhofer, J. | | Deposit date: | 1996-11-08 | | Release date: | 1997-11-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the bifunctional enzyme 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase reveals distinct domain homologies.
Structure, 4, 1996
|
|
1BHX
 
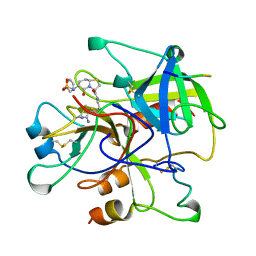 | |
1BH9
 
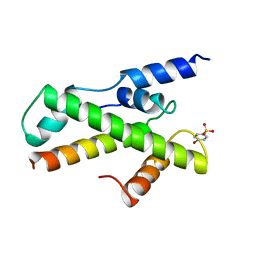 | | HTAFII18/HTAFII28 HETERODIMER CRYSTAL STRUCTURE WITH BOUND PCMBS | | Descriptor: | PARA-MERCURY-BENZENESULFONIC ACID, TAFII18, TAFII28 | | Authors: | Birck, C, Poch, O, Romier, C, Ruff, M, Mengus, G, Lavigne, A.-C, Davidson, I, Moras, D. | | Deposit date: | 1998-06-16 | | Release date: | 1999-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human TAF(II)28 and TAF(II)18 interact through a histone fold encoded by atypical evolutionary conserved motifs also found in the SPT3 family.
Cell(Cambridge,Mass.), 94, 1998
|
|
1BHG
 
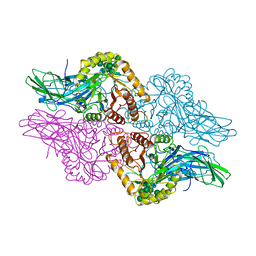 | | HUMAN BETA-GLUCURONIDASE AT 2.6 A RESOLUTION | | Descriptor: | BETA-GLUCURONIDASE, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Jain, S, Drendel, W.B. | | Deposit date: | 1996-03-04 | | Release date: | 1997-09-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structure of human beta-glucuronidase reveals candidate lysosomal targeting and active-site motifs.
Nat.Struct.Biol., 3, 1996
|
|
1BGJ
 
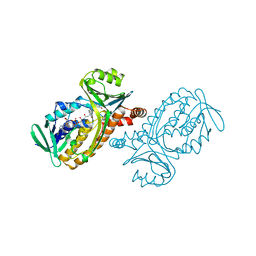 | | P-HYDROXYBENZOATE HYDROXYLASE (PHBH) MUTANT WITH CYS 116 REPLACED BY SER (C116S) AND HIS 162 REPLACED BY ARG (H162R), IN COMPLEX WITH FAD AND 4-HYDROXYBENZOIC ACID | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE, P-HYDROXYBENZOIC ACID | | Authors: | Eppink, M.H.M, Schreuder, H.A, Van Berkel, W.J.H. | | Deposit date: | 1998-05-29 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Interdomain binding of NADPH in p-hydroxybenzoate hydroxylase as suggested by kinetic, crystallographic and modeling studies of histidine 162 and arginine 269 variants.
J.Biol.Chem., 273, 1998
|
|
1BIO
 
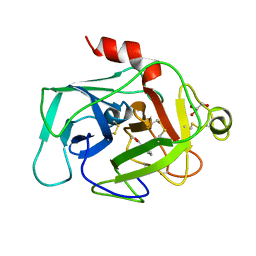 | | HUMAN COMPLEMENT FACTOR D IN COMPLEX WITH ISATOIC ANHYDRIDE INHIBITOR | | Descriptor: | COMPLEMENT FACTOR D, GLYCEROL, ISATOIC ANHYDRIDE | | Authors: | Jing, H, Babu, Y.S, Moore, D, Kilpatrick, J.M, Liu, X.-Y, Volanakis, J.E, Narayana, S.V.L. | | Deposit date: | 1998-06-18 | | Release date: | 1999-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of native and complexed complement factor D: implications of the atypical His57 conformation and self-inhibitory loop in the regulation of specific serine protease activity.
J.Mol.Biol., 282, 1998
|
|
1BAZ
 
 | |
1BHH
 
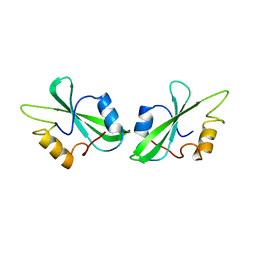 | | FREE P56LCK SH2 DOMAIN | | Descriptor: | P56 LCK TYROSINE KINASE SH2 DOMAIN, T-LYMPHOCYTE-SPECIFIC PROTEIN TYROSINE KINASE P56LCK | | Authors: | Tong, L, Warren, T.C, Lukas, S, Schembri-King, J, Betageri, R, Proudfoot, J.R, Jakes, S. | | Deposit date: | 1998-06-08 | | Release date: | 1998-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Carboxymethyl-phenylalanine as a replacement for phosphotyrosine in SH2 domain binding.
J.Biol.Chem., 273, 1998
|
|
1BIN
 
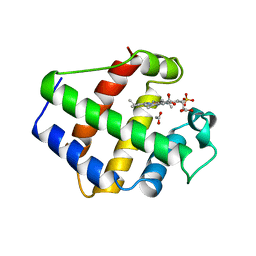 | | LEGHEMOGLOBIN A (ACETOMET) | | Descriptor: | ACETATE ION, LEGHEMOGLOBIN A, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Brucker, E.A, Hargrove, M.S, Phillips Jr, G.N. | | Deposit date: | 1996-08-23 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Characterization of recombinant soybean leghemoglobin a and apolar distal histidine mutants.
J.Mol.Biol., 266, 1997
|
|
1BIS
 
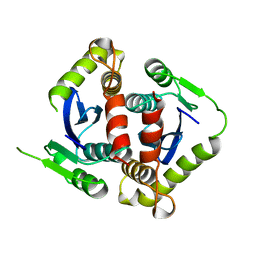 | | HIV-1 INTEGRASE CORE DOMAIN | | Descriptor: | HIV-1 INTEGRASE | | Authors: | Goldgur, Y, Dyda, F, Hickman, A.B, Jenkins, T.M, Craigie, R, Davies, D.R. | | Deposit date: | 1998-06-19 | | Release date: | 1998-08-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1BGW
 
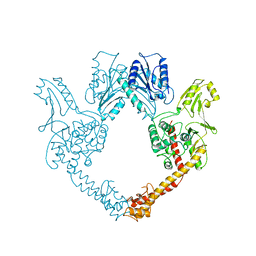 | | TOPOISOMERASE RESIDUES 410-1202, | | Descriptor: | TOPOISOMERASE | | Authors: | Berger, J.M, Gamblin, S.J, Harrison, S.C, Wang, J.C. | | Deposit date: | 1996-02-20 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and mechanism of DNA topoisomerase II.
Nature, 379, 1996
|
|
