4HMP
 
 | | Crystal structure of iron uptake ABC transporter substrate-binding protein PiaA from Streptococcus pneumoniae TIGR4 | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, GLYCEROL, ... | | Authors: | Cheng, W, Li, Q, Jiang, Y.-L, Chen, Y, Zhou, C.-Z. | | Deposit date: | 2012-10-18 | | Release date: | 2013-09-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of Streptococcus pneumoniae PiaA and Its Complex with Ferrichrome Reveal Insights into the Substrate Binding and Release of High Affinity Iron Transporters
Plos One, 8, 2013
|
|
5LA4
 
 | | Crystal structure of apo human proheparanase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Heparanase, ... | | Authors: | Wu, L, Jin, Y, Davies, G.J. | | Deposit date: | 2016-06-13 | | Release date: | 2017-05-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Activity-based probes for functional interrogation of retaining beta-glucuronidases.
Nat. Chem. Biol., 13, 2017
|
|
3KKA
 
 | | Co-crystal structure of the sam domains of EPHA1 AND EPHA2 | | Descriptor: | CHLORIDE ION, EPHRIN TYPE-A RECEPTOR 1, EPHRIN TYPE-A RECEPTOR 2 | | Authors: | Walker, J.R, Yermekbayeva, L, Butler-Cole, C, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-11-05 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Co-Crystal Structure of the SAM Domains of Human Ephrin Type-A Receptor 1 and Human Ephrin Type-A Receptor 2
To be Published
|
|
4HTU
 
 | | Structure of 5-chlorouracil modified A:U base pair | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*(UCL)P*CP*GP*CP*G)-3'), GLYCEROL, ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2012-11-01 | | Release date: | 2012-12-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.489 Å) | | Cite: | Structure, stability and function of 5-chlorouracil modified A:U and G:U base pairs.
Nucleic Acids Res., 41, 2013
|
|
5EU9
 
 | | Structure of Human Enolase 2 in complex with ((3S,5S)-1,5-dihydroxy-3-methyl-2-oxopyrrolidin-3-yl)phosphonic acid | | Descriptor: | ((3S,5S)-1,5-dihydroxy-3-methyl-2-oxopyrrolidin-3-yl)phosphonic acid, Gamma-enolase, MAGNESIUM ION, ... | | Authors: | Leonard, P.G, Muller, F.L. | | Deposit date: | 2015-11-18 | | Release date: | 2016-11-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | SF2312, a natural phosphonate inhibitor of Enolase
To be Published
|
|
2Q4T
 
 | | Ensemble refinement of the protein crystal structure of a cytosolic 5'-nucleotidase III from Mus musculus Mm.158936 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Cytosolic 5'-nucleotidase III | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
3AHW
 
 | |
6P1L
 
 | | Crystal structure of EGFR in complex with EAI045 | | Descriptor: | (2R)-2-(5-fluoro-2-hydroxyphenyl)-2-(1-oxo-1,3-dihydro-2H-isoindol-2-yl)-N-(1,3-thiazol-2-yl)acetamide, Epidermal growth factor receptor, MAGNESIUM ION, ... | | Authors: | Heppner, D.E, Eck, M.J. | | Deposit date: | 2019-05-20 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and Optimization of Dibenzodiazepinones as Allosteric Mutant-Selective EGFR Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
1J53
 
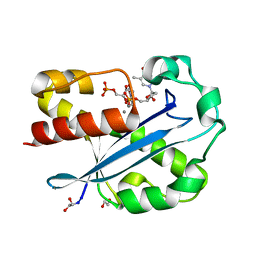 | | Structure of the N-terminal Exonuclease Domain of the Epsilon Subunit of E.coli DNA Polymerase III at pH 8.5 | | Descriptor: | 1,2-ETHANEDIOL, DNA polymerase III, epsilon chain, ... | | Authors: | Hamdan, S, Carr, P.D, Brown, S.E, Ollis, D.L, Dixon, N.E. | | Deposit date: | 2002-01-22 | | Release date: | 2002-10-16 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Proofreading during Replication of the Escherichia coli Chromosome
Structure, 10, 2002
|
|
5ESS
 
 | | Crystal Structure of M. tuberculosis MenD bound to Mg2+ and covalent intermediate I (a ThDP and decarboxylated 2-oxoglutarate adduct) | | Descriptor: | 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase, 4-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-5-[2-[oxidanyl(phosphonooxy)phosphoryl]oxyethyl]-1,3-thiazol-3 -ium-2-yl]-4-oxidanyl-butanoic acid, ACETATE ION, ... | | Authors: | Johnston, J.M, Jirgis, E.N.M, Bashiri, G, Bulloch, E.M.M, Baker, E.N. | | Deposit date: | 2015-11-17 | | Release date: | 2016-06-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Views along the Mycobacterium tuberculosis MenD Reaction Pathway Illuminate Key Aspects of Thiamin Diphosphate-Dependent Enzyme Mechanisms.
Structure, 24, 2016
|
|
3KDN
 
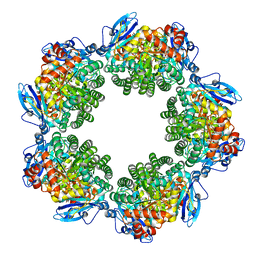 | | Crystal structure of Type III Rubisco SP4 mutant complexed with 2-CABP | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Nishitani, Y, Fujihashi, M, Doi, T, Yoshida, S, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2009-10-23 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure-based catalytic optimization of a type III Rubisco from a hyperthermophile
J.Biol.Chem., 285, 2010
|
|
4XUG
 
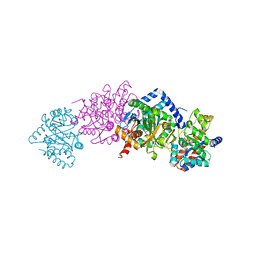 | | Crystal structure of Tryptophan Synthase from Salmonella typhimurium in complex with 2-({[4-(Trifluoromethoxy)Phenyl]Sulfonyl}Amino)Ethyl Dihydrogen Phosphate (F9F) inhibitor in the alpha site and ammonium ion in the metal coordination site. | | Descriptor: | 1,2-ETHANEDIOL, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, AMMONIUM ION, ... | | Authors: | Hilario, E, Caulkins, B.G, Young, R.P, Niks, D, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2015-01-25 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of Tryptophan Synthase from Salmonella typhimurium in complex with 2-({[4-(Trifluoromethoxy)Phenyl]Sulfonyl}Amino)Ethyl Dihydrogen Phosphate (F9F) inhibitor in the alpha site and ammonium ion in the metal coordination site.
To Be Published
|
|
4LWX
 
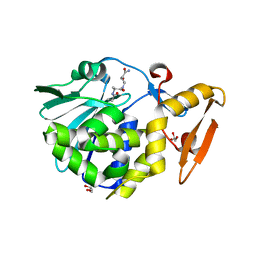 | | Crystal structure of the complex of Ribosome inactivating protein from Momordica Balsamina with peptidoglycan fragment at 1.78 A resolution | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ALANINE, ... | | Authors: | Yamini, S, Pandey, S, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-07-29 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of the complex of Ribosome inactivating protein from Momordica Balsamina with peptidoglycan fragment at 1.78 A resolution
To be Published
|
|
4XUX
 
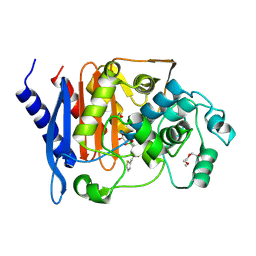 | | Structure of ampC bound to RPX-7009 at 1.75 A | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, Beta-lactamase, ... | | Authors: | Clifton, M.C, Abendroth, J. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of a Cyclic Boronic Acid beta-Lactamase Inhibitor (RPX7009) with Utility vs Class A Serine Carbapenemases.
J.Med.Chem., 58, 2015
|
|
6DA3
 
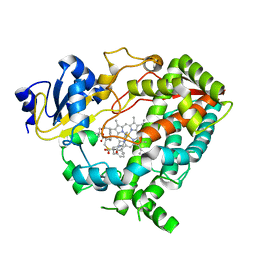 | | Human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl [(2R)-1-{[(2R)-1-oxo-3-phenyl-1-{[(pyridin-3-yl)methyl]amino}propan-2-yl]sulfanyl}-3-phenylpropan-2-yl]carbamate | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2018-05-01 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure-Activity Relationships of Rationally Designed Ritonavir Analogues: Impact of Side-Group Stereochemistry, Headgroup Spacing, and Backbone Composition on the Interaction with CYP3A4.
Biochemistry, 58, 2019
|
|
4HU1
 
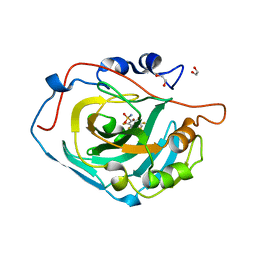 | | Crystal structure of human carbonic anhydrase isozyme XIII with the inhibitor. | | Descriptor: | 1,2-ETHANEDIOL, 2,3,5,6-tetrafluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, Carbonic anhydrase 13, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2012-11-02 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 4-Substituted-2,3,5,6-tetrafluorobenzenesulfonamides as inhibitors of carbonic anhydrases I, II, VII, XII, and XIII.
Bioorg.Med.Chem., 21, 2013
|
|
4HN0
 
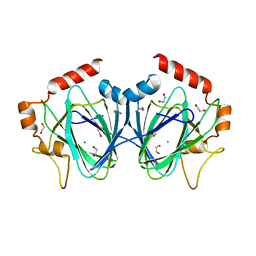 | |
8GF2
 
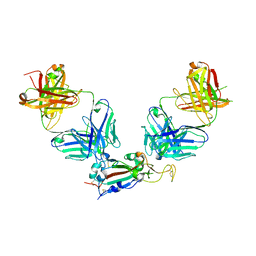 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with antibodies eCR3022.20 and CC12.3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CC12.3 Fab heavy chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-03-07 | | Release date: | 2023-08-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | Broadening a SARS-CoV-1-neutralizing antibody for potent SARS-CoV-2 neutralization through directed evolution.
Sci.Signal., 16, 2023
|
|
5X8M
 
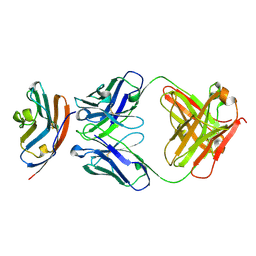 | | PD-L1 in complex with durvalumab | | Descriptor: | Programmed cell death 1 ligand 1, durvalumab heavy chain, durvalumab light chain | | Authors: | Heo, Y.S, Lee, H.T. | | Deposit date: | 2017-03-03 | | Release date: | 2017-08-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.661 Å) | | Cite: | Molecular mechanism of PD-1/PD-L1 blockade via anti-PD-L1 antibodies atezolizumab and durvalumab
Sci Rep, 7, 2017
|
|
8GO3
 
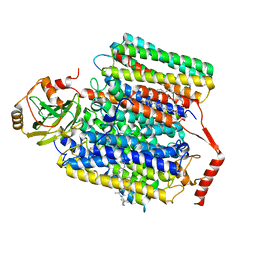 | |
6DGK
 
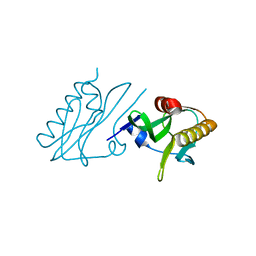 | |
9BJ1
 
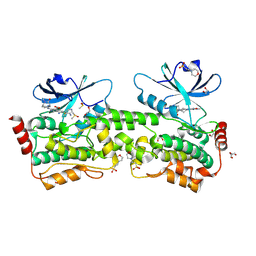 | | Crystal structure of inhibitor GNE-6893 bound to HPK1 | | Descriptor: | (4S,5R,7R,11aP)-10-{[(3R)-3-hydroxy-1-methyl-2-oxopyrrolidin-3-yl]ethynyl}-N~3~-methyl-6,7-dihydro-5H-5,7-methanoimidazo[2,1-a][2]benzazepine-2,3-dicarboxamide, (9S)-2-{[(6P)-8-amino-6-(5-amino-4-methylpyridin-3-yl)-7-fluoroisoquinolin-3-yl]amino}-6-methyl-5,6-dihydro-4H-pyrazolo[1,5-d][1,4]diazepin-7(8H)-one, 1,2-ETHANEDIOL, ... | | Authors: | Kiefer, J.R, Tellis, J.C, Chan, B.K, Wang, W, Wu, P, Choo, E.F, Heffron, T.P, Wei, B, Siu, M. | | Deposit date: | 2024-04-24 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Discovery of GNE-6893, a Potent, Selective, Orally Bioavailable Small Molecule Inhibitor of HPK1.
Acs Med.Chem.Lett., 15, 2024
|
|
5F2I
 
 | |
5SQ1
 
 | |
5BTG
 
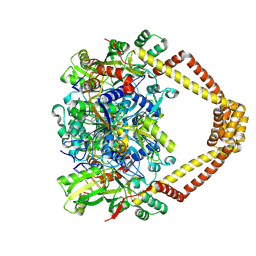 | | Crystal structure of a topoisomerase II complex | | Descriptor: | (3S)-9-fluoro-3-methyl-10-(4-methylpiperazin-1-yl)-7-oxo-2,3-dihydro-7H-[1,4]oxazino[2,3,4-ij]quinoline-6-carboxylic acid, DNA gyrase subunit A, DNA gyrase subunit B, ... | | Authors: | Blower, T.R, Williamson, B.H, Kerns, R.J, Berger, J.M. | | Deposit date: | 2015-06-03 | | Release date: | 2016-03-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and stability of gyrase-fluoroquinolone cleaved complexes from Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
