4NA8
 
 | | Factor XIa in complex with the inhibitor 5-aminocarbonyl-2-[3-[(2s,4r)-6-carbamimidoyl-4-methyl-4-phenyl-2,3-dihydro-1h-quinolin-2-yl]phenyl]benzoic acid | | Descriptor: | 1,2-ETHANEDIOL, 5-aminocarbonyl-2-[3-[(2S,4R)-6-carbamimidoyl-4-methyl-4-phenyl-2,3-dihydro-1H-quinolin-2-yl]phenyl]benzoic acid, Coagulation factor XI, ... | | Authors: | Wei, A. | | Deposit date: | 2013-10-21 | | Release date: | 2014-02-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tetrahydroquinoline Derivatives as Potent and Selective Factor XIa Inhibitors.
J.Med.Chem., 57, 2014
|
|
5AXW
 
 | | Crystal structure of Staphylococcus aureus Cas9 in complex with sgRNA and target DNA (TTGGGT PAM) | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, DNA (28-MER), ... | | Authors: | Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2015-08-01 | | Release date: | 2015-09-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Staphylococcus aureus Cas9.
Cell, 162, 2015
|
|
5ENC
 
 | | Crystal structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with N-(2,6-Dichlorobenzyl)acetamide (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, PH-interacting protein, ~{N}-[[2,6-bis(chloranyl)phenyl]methyl]ethanamide | | Authors: | Krojer, T, Talon, R, Collins, P, Bradley, A, Cox, O, Szykowska, A, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | A poised fragment library enables rapid synthetic expansion yielding the first reported inhibitors of PHIP(2), an atypical bromodomain.
Chem Sci, 7, 2016
|
|
5EKT
 
 | |
7LTL
 
 | |
5VMK
 
 | |
5B68
 
 | | Crystal structure of apo amylomaltase from Corynebacterium glutamicum | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-alpha-glucanotransferase, GLYCEROL, ... | | Authors: | Joo, S, Kim, S, Kim, K.-J. | | Deposit date: | 2016-05-25 | | Release date: | 2016-08-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Amylomaltase from Corynebacterium glutamicum.
J.Agric.Food Chem., 64, 2016
|
|
7LK3
 
 | | Crystal structure of untwinned human GABARAPL2 | | Descriptor: | 1,2-ETHANEDIOL, Gamma-aminobutyric acid receptor-associated protein-like 2 | | Authors: | Scicluna, K, Dewson, G, Czabotar, P.E, Birkinshaw, R.W. | | Deposit date: | 2021-02-01 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A new crystal form of GABARAPL2.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
3M11
 
 | | Crystal Structure of Aurora A Kinase complexed with inhibitor | | Descriptor: | 1-(4-{2-[(5,6-diphenylfuro[2,3-d]pyrimidin-4-yl)amino]ethyl}phenyl)-3-phenylurea, Serine/threonine-protein kinase 6 | | Authors: | Wu, J.S, Leou, J.S, Coumar, M.S, Hsieh, H.P, Wu, S.Y. | | Deposit date: | 2010-03-03 | | Release date: | 2011-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Fast-forwarding hit to lead: aurora and epidermal growth factor receptor kinase inhibitor lead identification
J.Med.Chem., 53, 2010
|
|
3M1G
 
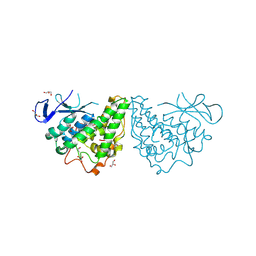 | | The structure of a putative glutathione S-transferase from Corynebacterium glutamicum | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Putative glutathione S-transferase | | Authors: | Cuff, M.E, Marshall, N, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-04 | | Release date: | 2010-04-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of a putative glutathione S-transferase from Corynebacterium glutamicum
TO BE PUBLISHED
|
|
2YPV
 
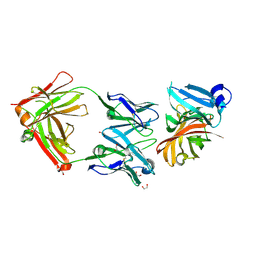 | | Crystal structure of the Meningococcal vaccine antigen factor H binding protein in complex with a bactericidal antibody | | Descriptor: | 1,2-ETHANEDIOL, FAB 12C1, LIPOPROTEIN | | Authors: | Malito, E, Veggi, D, Bottomley, M.J. | | Deposit date: | 2012-11-01 | | Release date: | 2013-02-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Defining a Protective Epitope on Factor H Binding Protein, a Key Meningococcal Virulence Factor and Vaccine Antigen.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6L07
 
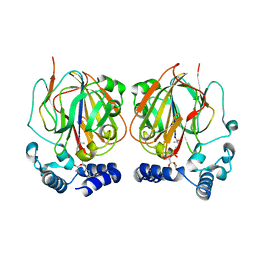 | |
4JO5
 
 | | CBM3a-L domain with flanking linkers from scaffoldin cipA of cellulosome of Clostridium thermocellum | | Descriptor: | 1-METHYLIMIDAZOLE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Shimon, L.J.W, Frolow, F, Bayer, E.A, Yaniv, O, Lamed, R, Morag, E. | | Deposit date: | 2013-03-16 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of a family 3a carbohydrate-binding module from the cellulosomal scaffoldin CipA of Clostridium thermocellum with flanking linkers: implications for cellulosome structure.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4NJD
 
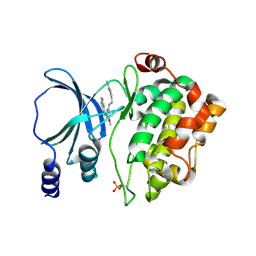 | | Structure of p21-activated kinase 4 with a novel inhibitor KY-04031 | | Descriptor: | N-(1H-indazol-5-yl)-N'-[2-(1H-indol-3-yl)ethyl]-6-methoxy-1,3,5-triazine-2,4-diamine, Serine/threonine-protein kinase PAK 4 | | Authors: | Park, S. | | Deposit date: | 2013-11-09 | | Release date: | 2014-05-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery and the structural basis of a novel p21-activated kinase 4 inhibitor.
Cancer Lett., 349, 2014
|
|
7KYD
 
 | | Drosophila melanogaster long-chain fatty-acyl-CoA synthetase CG6178 | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-[(S)-hydroxy(octanoyloxy)phosphoryl]adenosine, Long-chain fatty-acyl-CoA synthetase CG6178 | | Authors: | Adams, S.T, Zephyr, J, Bohn, M.F, Schiffer, C.A, Miller, S.C. | | Deposit date: | 2020-12-07 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | FruitFire: a luciferase based on a fruit fly metabolic enzyme.
Biorxiv, 2023
|
|
4L3O
 
 | | Crystal Structure of SIRT2 in complex with the macrocyclic peptide S2iL5 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Yamagata, K, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2013-06-06 | | Release date: | 2014-02-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.518 Å) | | Cite: | Structural Basis for Potent Inhibition of SIRT2 Deacetylase by a Macrocyclic Peptide Inducing Dynamic Structural Change
Structure, 22, 2013
|
|
1D40
 
 | | BASE SPECIFIC BINDING OF COPPER(II) TO Z-DNA: THE 1.3-ANGSTROMS SINGLE CRYSTAL STRUCTURE OF D(M5CGUAM5CG) IN THE PRESENCE OF CUCL2 | | Descriptor: | COPPER (II) CHLORIDE, COPPER (II) ION, DNA (5'-D(*(5CM)P*(CU)GP*UP*AP*(5CM)P*(CU)G)-3') | | Authors: | Geierstanger, B.H, Kagawa, T.F, Chen, S.-L, Quigley, G.J, Ho, P.S. | | Deposit date: | 1991-05-07 | | Release date: | 1992-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Base-specific binding of copper(II) to Z-DNA. The 1.3-A single crystal structure of d(m5CGUAm5CG) in the presence of CuCl2.
J.Biol.Chem., 266, 1991
|
|
4BCR
 
 | | Structure of PPARalpha in complex with WY14643 | | Descriptor: | 1,2-ETHANEDIOL, 2-({4-CHLORO-6-[(2,3-DIMETHYLPHENYL)AMINO]PYRIMIDIN-2-YL}SULFANYL)ACETIC ACID, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR ALPHA | | Authors: | Bernardes, A, Muniz, J.R.C, Polikarpov, I. | | Deposit date: | 2012-10-02 | | Release date: | 2013-05-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Molecular Mechanism of Peroxisome Proliferator-Activated Receptor Alpha Activation by Wy14643: A New Mode of Ligand Recognition and Receptor Stabilization
J.Mol.Biol., 425, 2013
|
|
2ZWA
 
 | | Crystal structure of tRNA wybutosine synthesizing enzyme TYW4 | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, Leucine carboxyl methyltransferase 2, ... | | Authors: | Suzuki, Y, Noma, A, Suzuki, T, Ishitani, R, Nureki, O. | | Deposit date: | 2008-12-01 | | Release date: | 2009-06-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of tRNA modification with CO2 fixation and methylation by wybutosine synthesizing enzyme TYW4.
Nucleic Acids Res., 37, 2009
|
|
3E0Q
 
 | | Crystal structure of Schistosoma mansoni purine nucleoside phosphorylase complexed with a novel monocyclic inhibitor | | Descriptor: | 6-amino-5-bromo-1,2,3,4-tetrahydropyrimidine-2,4-dione, DIMETHYL SULFOXIDE, Purine-nucleoside phosphorylase | | Authors: | Pereira, H.M, Berdini, V, Ferri, M.R, Cleasby, A, Garratt, R.C. | | Deposit date: | 2008-07-31 | | Release date: | 2009-08-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Schistosoma purine nucleoside phosphorylase complexed with a novel monocyclic inhibitor.
Acta Trop., 114, 2010
|
|
3DWY
 
 | | Crystal Structure of the Bromodomain of Human CREBBP | | Descriptor: | 1,2-ETHANEDIOL, CREB-binding protein | | Authors: | Filippakopoulos, P, Picaud, S, Fedorov, O, Karim, R, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Wickstroem, M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-07-23 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
3EBU
 
 | | Replacement of Val3 in Human Thymidylate Synthase Affects Its Kinetic Properties and Intracellular Stability | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Thymidylate synthase | | Authors: | Huang, X, Gibson, L.M, Bell, B.J, Lovelace, L.L, Pena, M.M, Berger, F.G, Berger, S.H. | | Deposit date: | 2008-08-28 | | Release date: | 2010-03-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Replacement of Val3 in human thymidylate synthase affects its kinetic properties and intracellular stability .
Biochemistry, 49, 2010
|
|
2K1Y
 
 | | Solution Structure of Duplex DNA Containing the Mutagenic Lesion: 1,N2-Etheno-2'-deoxyguanine | | Descriptor: | 5'-D(*DCP*DGP*DTP*DAP*DCP*(GNE)P*DCP*DAP*DTP*DGP*DC)-3', 5'-D(*DGP*DCP*DAP*DTP*DGP*DCP*DGP*DTP*DAP*DCP*DG)-3' | | Authors: | Zaliznyak, T, Lukin, M, Johnson, F, de los Santos, C. | | Deposit date: | 2008-03-18 | | Release date: | 2009-02-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of duplex DNA containing the mutagenic lesion 1,N(2)-etheno-2'-deoxyguanine.
Biochemistry, 47, 2008
|
|
1DMF
 
 | | THE THREE-DIMENSIONAL SOLUTION STRUCTURE OF CALLINECTES SAPIDUS METALLOTHIONEIN-I DETERMINED BY HOMONUCLEAR AND HETERONUCLEAR MAGNETIC RESONANCE SPECTOSCOPY | | Descriptor: | CADMIUM ION, CD6 METALLOTHIONEIN-1 | | Authors: | Narula, S.S, Brouwer, M, Hua, Y, Armitage, I.M. | | Deposit date: | 1994-11-22 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of Callinectes sapidus metallothionein-1 determined by homonuclear and heteronuclear magnetic resonance spectroscopy.
Biochemistry, 34, 1995
|
|
1MKI
 
 | | Crystal Structure of Bacillus Subtilis Probable Glutaminase, APC1040 | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Probable Glutaminase ybgJ | | Authors: | Kim, Y, Dementieva, I, Vinokour, E, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-08-29 | | Release date: | 2003-06-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and structural characterization of four glutaminases from Escherichia coli and Bacillus subtilis.
Biochemistry, 47, 2008
|
|
