5DKY
 
 | | Crystal structure of glucosidase II alpha subunit (DNJ-bound from) | | Descriptor: | 1-DEOXYNOJIRIMYCIN, Alpha glucosidase-like protein | | Authors: | Satoh, T, Toshimori, T, Yan, G, Yamaguchi, T, Kato, K. | | Deposit date: | 2015-09-04 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for two-step glucose trimming by glucosidase II involved in ER glycoprotein quality control.
Sci Rep, 6, 2016
|
|
5V7C
 
 | |
6SZF
 
 | | Solution structure of the amyloid beta-peptide (1-42) | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Grimaldi, M, Santoro, A, Stillitano, I, Buonocore, M, D'Ursi, A.M. | | Deposit date: | 2019-10-02 | | Release date: | 2020-10-14 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Exploring the Early Stages of the Amyloid A beta (1-42) Peptide Aggregation Process: An NMR Study.
Pharmaceuticals, 14, 2021
|
|
4ZXC
 
 | | Crystal Structure of hydroquinone 1,2-dioxygenase PnpCD in complex with Fe3+ | | Descriptor: | FE (III) ION, Hydroquinone dioxygenase large subunit, Hydroquinone dioxygenase small subunit | | Authors: | Liu, S, Su, T, Zhang, C, Gu, L. | | Deposit date: | 2015-05-20 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal Structure of PnpCD, a Two-subunit Hydroquinone 1,2-Dioxygenase, Reveals a Novel Structural Class of Fe2+-dependent Dioxygenases.
J.Biol.Chem., 290, 2015
|
|
7F1X
 
 | | X-ray crystal structure of visual arrestin complexed with inositol 1,4,5-triphosphate | | Descriptor: | 1,2-ETHANEDIOL, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, PENTANEDIAL, ... | | Authors: | Jang, K, Kang, M, Eger, B.T, Choe, H.W, Ernst, O.P, Kim, Y.J. | | Deposit date: | 2021-06-10 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural evidence for visual arrestin priming via complexation of phosphoinositols.
Structure, 30, 2022
|
|
8E73
 
 | | Vigna radiata supercomplex I+III2 (full bridge) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | Authors: | Maldonado, M, Letts, J.A. | | Deposit date: | 2022-08-23 | | Release date: | 2023-01-11 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Plant-specific features of respiratory supercomplex I + III 2 from Vigna radiata.
Nat.Plants, 9, 2023
|
|
2RIZ
 
 | |
5UIO
 
 | |
9DWI
 
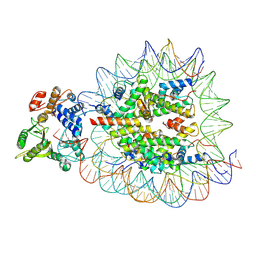 | | DNA Polymerase Beta bound to a nucleosome containing a 1-nt gap at SHL-4.5 (State 3, composite) | | Descriptor: | 601 I strand (damaged strand 1), 601 J strand (non-damaged strand), 601 K strand (damaged strand 2), ... | | Authors: | Weaver, T.M, Ryan, B.J, Freudenthal, B.D. | | Deposit date: | 2024-10-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | DNA Polymerase Beta bound to a nucleosome containing a 1-nt gap at SHL-4.5 (State 4, composite)
To Be Published
|
|
5CBU
 
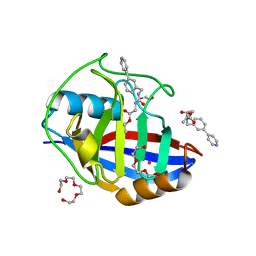 | | Human Cyclophilin D Complexed with Inhibitor. | | Descriptor: | 1,2-ETHANEDIOL, POTASSIUM ION, Peptidyl-prolyl cis-trans isomerase F, ... | | Authors: | Gibson, R.P, Shore, E, Kershaw, N, Awais, M, Javed, A, Latawiec, D, Pandalaneni, S, Wen, L, Berry, N, O'Neill, P, Sutton, R, Lian, L.Y. | | Deposit date: | 2015-07-01 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Human Cyclophilin D Complexed with Inhibitor.
To Be Published
|
|
6QN4
 
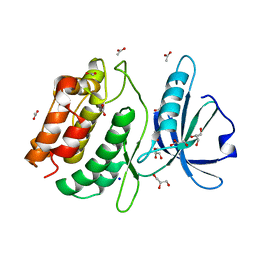 | |
6QOD
 
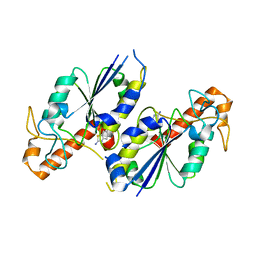 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with Fragment 7 (2-Amino-6-fluorobenzothiazole) | | Descriptor: | 6-fluoro-1,3-benzothiazol-2-amine, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-12 | | Release date: | 2020-03-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Fragment-based discovery of a new class of inhibitors targeting mycobacterial tRNA modification.
Nucleic Acids Res., 48, 2020
|
|
6FQN
 
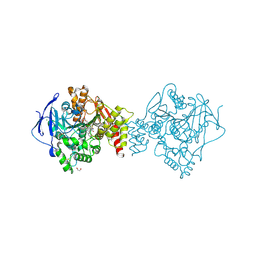 | | Carbamylated T. californica acetylcholineterase bound to uncharged hybrid reactivator 2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-[4-[(7-chloranyl-1,2,3,4-tetrahydroacridin-9-yl)amino]butyl]-2-[(oxidanylamino)methyl]pyridin-3-ol, ... | | Authors: | De la Mora, E, Santoni, G, de Souza, J, Sussman, J, Silman, I, Baati, R, Weik, M, Nachon, F. | | Deposit date: | 2018-02-14 | | Release date: | 2018-08-29 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.30002666 Å) | | Cite: | Structure-Based Optimization of Nonquaternary Reactivators of Acetylcholinesterase Inhibited by Organophosphorus Nerve Agents.
J. Med. Chem., 61, 2018
|
|
8GFV
 
 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #1 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
5C0C
 
 | | 1E6 TCR in complex with HLA-A02 carrying RQFGPDWIVA | | Descriptor: | 1,2-ETHANEDIOL, 1E6 TCR Alpha Chain, 1E6 TCR Beta Chain, ... | | Authors: | Rizkallah, P.J, Bulek, A.M, Cole, D.K, Sewell, A.K. | | Deposit date: | 2015-06-12 | | Release date: | 2016-05-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.974 Å) | | Cite: | Hotspot autoimmune T cell receptor binding underlies pathogen and insulin peptide cross-reactivity.
J.Clin.Invest., 126, 2016
|
|
7S1Q
 
 | | PRMT5/MEP50 crystal structure with MTA and a phthalazinone inhibitor bound (Compound 9) | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, Methylosome protein 50, Protein arginine N-methyltransferase 5, ... | | Authors: | Gunn, R.J, Thomas, N.C, Lawson, J.D, Ivetac, A, Smith, C.R, Kulyk, S, Marx, M.A. | | Deposit date: | 2021-09-02 | | Release date: | 2022-01-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Fragment-Based Discovery of MRTX1719, a Synthetic Lethal Inhibitor of the PRMT5•MTA Complex for the Treatment of MTAP -Deleted Cancers.
J.Med.Chem., 65, 2022
|
|
6FSY
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 3,5-dimethylisoxazol ligand | | Descriptor: | 1,2-ETHANEDIOL, 3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-[(~{R})-oxidanyl(pyridin-3-yl)methyl]phenol, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Conway, S.J, Pike, A.C.W, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-20 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | BET bromodomain ligands: Probing the WPF shelf to improve BRD4 bromodomain affinity and metabolic stability.
Bioorg.Med.Chem., 26, 2018
|
|
9DWM
 
 | |
6FT7
 
 | | Crystal structure of CLK3 in complex with compound 8a | | Descriptor: | 1,2-ETHANEDIOL, 3-phenyl-1~{H}-pyrrolo[3,4-g]indol-8-one, Dual specificity protein kinase CLK3, ... | | Authors: | Chaikuad, A, Walter, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Kunick, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-20 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Molecular structures of cdc2-like kinases in complex with a new inhibitor chemotype.
PLoS ONE, 13, 2018
|
|
6FTM
 
 | | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-048 | | Descriptor: | 1,2-ETHANEDIOL, 3-{5-[(4aR,8aS)-3-cycloheptyl-4-oxo-3,4,4a,5,8,8a-hexahydrophthalazin-1-yl]-2-methoxyphenyl}prop-2-ynamide, FORMIC ACID, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2018-02-22 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-048
To be published
|
|
9DWG
 
 | | DNA Polymerase Beta bound to a nucleosome containing a 1-nt gap at SHL-4.5 (State 1, composite) | | Descriptor: | 601 I strand (damaged strand 1), 601 J strand (non-damaged strand), 601 K strand (damaged strand 2), ... | | Authors: | Weaver, T.M, Ryan, B.J, Freudenthal, B.D. | | Deposit date: | 2024-10-09 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | DNA Polymerase Beta bound to a nucleosome containing a 1-nt gap at SHL-4.5 (State 1, composite)
To Be Published
|
|
7S1S
 
 | | PRMT5/MEP50 crystal structure with MTA and MRTX-1719 bound | | Descriptor: | (7-{(5M)-5-[3-chloro-6-cyano-5-(cyclopropyloxy)-2-fluorophenyl]-1-methyl-1H-pyrazol-4-yl}-4-oxo-3,4-dihydrophthalazin-1-yl)methanaminium, 5'-DEOXY-5'-METHYLTHIOADENOSINE, Methylosome protein 50, ... | | Authors: | Gunn, R.J, Thomas, N.C, Lawson, J.D, Ivetac, A, Kulyk, S, Smith, C.R, Marx, M.A. | | Deposit date: | 2021-09-02 | | Release date: | 2022-01-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Fragment-Based Discovery of MRTX1719, a Synthetic Lethal Inhibitor of the PRMT5•MTA Complex for the Treatment of MTAP -Deleted Cancers.
J.Med.Chem., 65, 2022
|
|
5FOG
 
 | | Crystal structure of hte Cryptosporidium muris cytosolic leucyl-tRNA synthetase editing domain complex with a post-transfer editing analogue of norvaline (Nv2AA) | | Descriptor: | 1,2-ETHANEDIOL, 2'-(L-NORVALYL)AMINO-2'-DEOXYADENOSINE, LEUCYL-TRNA SYNTHETASE, ... | | Authors: | Palencia, A, Liu, R.J, Lukarska, M, Gut, J, Bougdour, A, Touquet, B, Wang, E.D, Alley, M.R.K, Rosenthal, P.J, Hakimi, M.A, Cusack, S. | | Deposit date: | 2015-11-20 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cryptosporidium and Toxoplasma Parasites are Inhibited by a Benzoxaborole Targeting Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
5C3F
 
 | | Crystal structure of Mcl-1 bound to BID-MM | | Descriptor: | BID-MM, GLYCEROL, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Miles, J.A, Yeo, D.J, Rowell, P, Rodriguez-Marin, S, Pask, C.M, Warriner, S.L, Edwards, T.A, Wilson, A.J. | | Deposit date: | 2015-06-17 | | Release date: | 2016-04-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Hydrocarbon constrained peptides - understanding preorganisation and binding affinity.
Chem Sci, 7, 2016
|
|
7Y5U
 
 | | Cryo-EM structure of the monomeric human CAF1LC-H3-H4 complex | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, Z.Y, Yu, C, Xu, R.M. | | Deposit date: | 2022-06-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
