4QT8
 
 | | Crystal Structure of RON Sema-PSI-IPT1 extracellular domains in complex with MSP beta-chain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hepatocyte growth factor-like protein, Macrophage-stimulating protein receptor, ... | | Authors: | Herzberg, O, Chao, K.L. | | Deposit date: | 2014-07-07 | | Release date: | 2014-09-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the binding specificity of human Recepteur d'Origine Nantais (RON) receptor tyrosine kinase to macrophage-stimulating protein.
J.Biol.Chem., 289, 2014
|
|
1DJ2
 
 | | STRUCTURES OF ADENYLOSUCCINATE SYNTHETASE FROM TRITICUM AESTIVUM AND ARABIDOPSIS THALIANA | | Descriptor: | ADENYLOSUCCINATE SYNTHETASE, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Prade, L, Cowan-Jacob, S.W, Chemla, P, Potter, S, Ward, E, Fonne-Pfister, R. | | Deposit date: | 1999-12-01 | | Release date: | 2000-03-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of adenylosuccinate synthetase from Triticum aestivum and Arabidopsis thaliana.
J.Mol.Biol., 296, 2000
|
|
3R8D
 
 | |
3QI6
 
 | |
3S1J
 
 | | Crystal structure of acetate-bound hell's gate globin I | | Descriptor: | ACETATE ION, Hemoglobin-like flavoprotein, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Teh, A.H, Saito, J.A, Baharuddin, A, Tuckerman, J.R, Newhouse, J.S, Kanbe, M, Newhouse, E.I, Rahim, R.A, Favier, F, Didierjean, C, Sousa, E.H.S, Stott, M.B, Dunfield, P.F, Gonzalez, G, Gilles-Gonzalez, M.A, Najimudin, N, Alam, M. | | Deposit date: | 2011-05-15 | | Release date: | 2011-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hell's Gate globin I: an acid and thermostable bacterial hemoglobin resembling mammalian neuroglobin
Febs Lett., 585, 2011
|
|
2KXD
 
 | | The structure of SH3-F2 | | Descriptor: | 11-mer peptide,Spectrin alpha chain, non-erythrocytic 1,Spectrin alpha chain, non-erythrocytic 1 | | Authors: | Kutyshenko, V.P, Gushchina, L.V, Khristoforov, V.S, Prokhorov, D.A, Timchenko, M.A, Kudrevatykh, I.u.A, Fedyukina, D.V, Filimonov, V.V. | | Deposit date: | 2010-04-30 | | Release date: | 2011-04-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure and dynamics of the chimeric protein SH3-F2
Mol.Biol.(Engl.Transl.), 44, 2010
|
|
3S1I
 
 | | Crystal structure of oxygen-bound hell's gate globin I | | Descriptor: | Hemoglobin-like flavoprotein, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Teh, A.H, Saito, J.A, Baharuddin, A, Tuckerman, J.R, Newhouse, J.S, Kanbe, M, Newhouse, E.I, Rahim, R.A, Favier, F, Didierjean, C, Sousa, E.H.S, Stott, M.B, Dunfield, P.F, Gonzalez, G, Gilles-Gonzalez, M.A, Najimudin, N, Alam, M. | | Deposit date: | 2011-05-15 | | Release date: | 2011-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Hell's Gate globin I: an acid and thermostable bacterial hemoglobin resembling mammalian neuroglobin
Febs Lett., 585, 2011
|
|
2PKM
 
 | | Crystal structure of M tuberculosis Adenosine Kinase complexed with adenosine | | Descriptor: | ADENOSINE, Adenosine kinase | | Authors: | Reddy, M.C.M, Palaninathan, S.K, Shetty, N.D, Owen, J.L, Watson, M.D, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2007-04-17 | | Release date: | 2007-06-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High resolution crystal structures of Mycobacterium tuberculosis adenosine kinase: insights into the mechanism and specificity of this novel prokaryotic enzyme
J.Biol.Chem., 282, 2007
|
|
2DGJ
 
 | | Crystal structure of EbhA (756-1003 domain) from Staphylococcus aureus | | Descriptor: | ACETIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Tanaka, Y, Yao, M, Kuroda, M, Watanabe, N, Ohta, T, Tanaka, I. | | Deposit date: | 2006-03-14 | | Release date: | 2007-03-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A helical string of alternately connected three-helix bundles for the cell wall-associated adhesion protein Ebh from Staphylococcus aureus
Structure, 16, 2008
|
|
2NNQ
 
 | |
2L4K
 
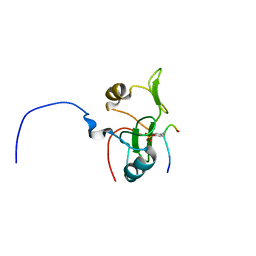 | | Water refined solution structure of the human Grb7-SH2 domain in complex with the 10 amino acid peptide pY1139 | | Descriptor: | Growth factor receptor-bound protein 7, Receptor tyrosine-protein kinase erbB-2 | | Authors: | Pias, S.C, Ivancic, M, Brescia, P.J, Johnson, D.L, Smith, D.E, Daly, R.J, Lyons, B.A. | | Deposit date: | 2010-10-07 | | Release date: | 2010-11-17 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Water-Refined Solution Structure of the Human Grb7-SH2 Domain in Complex with the erbB2 Receptor Peptide pY1139.
Protein Pept.Lett., 19, 2012
|
|
2OBS
 
 | |
2HXA
 
 | |
2HX9
 
 | |
2HX7
 
 | |
2HX8
 
 | |
2VN0
 
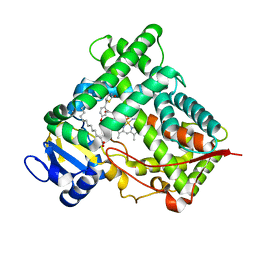 | | CYP2C8DH COMPLEXED WITH TROGLITAZONE | | Descriptor: | (5R)-5-(4-{[(2R)-6-HYDROXY-2,5,7,8-TETRAMETHYL-3,4-DIHYDRO-2H-CHROMEN-2-YL]METHOXY}BENZYL)-1,3-THIAZOLIDINE-2,4-DIONE, CYTOCHROME P450 2C8, PALMITIC ACID, ... | | Authors: | Schoch, G.A, Yano, J.K, Sansen, S, Stout, C.D, Johnson, E.F. | | Deposit date: | 2008-01-30 | | Release date: | 2008-04-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Determinants of cytochrome P450 2C8 substrate binding: structures of complexes with montelukast, troglitazone, felodipine, and 9-cis-retinoic acid.
J. Biol. Chem., 283, 2008
|
|
5PHE
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D in complex with N09453a | | Descriptor: | 1,2-ETHANEDIOL, 4-PHENYL-1H-IMIDAZOLE, Lysine-specific demethylase 4D, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.351 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5POA
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 in complex with N10186a | | Descriptor: | 1,2-ETHANEDIOL, 2-[5-(1H-pyrrol-1-yl)-1H-pyrazol-4-yl]pyridine, Bromodomain-containing protein 1, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PO4
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 in complex with N10170a | | Descriptor: | (4-nitrophenyl)methyl carbamimidothioate, 1,2-ETHANEDIOL, Bromodomain-containing protein 1, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.487 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
6LU9
 
 | |
5POJ
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 in complex with N10941a | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, N-(1-benzylpiperidin-4-yl)acetamide, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5POL
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 in complex with N10971a | | Descriptor: | 1,2-ETHANEDIOL, 1-(6,7-dimethoxy-3,4-dihydroisoquinolin-2(1H)-yl)ethan-1-one, Bromodomain-containing protein 1, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
8P1H
 
 | | Crystal structure of the chimera of human 14-3-3 zeta and phosphorylated cytoplasmic loop fragment of the alpha7 acetylcholine receptor | | Descriptor: | 1,2-ETHANEDIOL, AZIDE ION, BENZOIC ACID, ... | | Authors: | Boyko, K.M, Kapitonova, A.A, Tugaeva, K.V, Varfolomeeva, L.A, Lyukmanova, E.N, Sluchanko, N.N. | | Deposit date: | 2023-05-12 | | Release date: | 2023-10-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure reveals canonical recognition of the phosphorylated cytoplasmic loop of human alpha7 nicotinic acetylcholine receptor by 14-3-3 protein.
Biochem.Biophys.Res.Commun., 682, 2023
|
|
5POY
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 in complex with N11029a | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, N-[2-(phenylamino)ethyl]acetamide, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.758 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
