9M56
 
 | | Bovine Heart Cytochrome c Oxidase in the Fully Reduced State | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, 1,2-ETHANEDIOL, ... | | Authors: | Muramoto, K, Shinzawa-Itoh, K. | | Deposit date: | 2025-03-05 | | Release date: | 2025-07-02 | | Last modified: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The binding sites of carbon dioxide, nitrous oxide, and xenon reveal a putative exhaust channel for bovine cytochrome c oxidase.
J.Biol.Chem., 301, 2025
|
|
7G1T
 
 | | Crystal Structure of human FABP4 in complex with 6-methyl-3-phenyl-1,2,4-triazin-5-ol | | Descriptor: | 6-methyl-3-phenyl-1,2,4-triazin-5-ol, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Brunner, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
8OX9
 
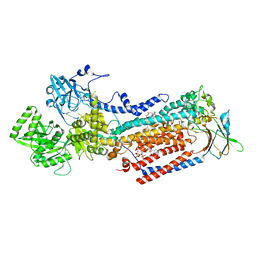 | | Cryo-EM structure of ATP8B1-CDC50A in E2P active conformation with bound PC | | Descriptor: | (2R)-3-{[(R)-{[(1S,2S,3R,4S,5S,6S)-2,6-dihydroxy-3,4,5-tris(phosphonooxy)cyclohexyl]oxy}(hydroxy)phosphoryl]oxy}propane -1,2-diyl dioctanoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dieudonne, T, Kummerer, F, Juknaviciute Laursen, M, Stock, C, Kock Flygaard, R, Khalid, S, Lenoir, G, Lyons, J.A, Lindorff-Larsen, K, Nissen, P. | | Deposit date: | 2023-05-01 | | Release date: | 2023-11-29 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Activation and substrate specificity of the human P4-ATPase ATP8B1.
Nat Commun, 14, 2023
|
|
7G1C
 
 | | Crystal Structure of human FABP4 in complex with rac-(1R,2S)-2-[(3-methylphenoxy)methyl]cyclohexane-1-carboxylic acid, i.e. SMILES C1CC[C@H]([C@H](C1)C(=O)O)COc1cc(ccc1)C with IC50=1.28108 microM | | Descriptor: | (1S,2R)-2-[(3-methylphenoxy)methyl]cyclohexane-1-carboxylic acid, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Buettelmann, B, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
7FYF
 
 | | Crystal Structure of human FABP4 in complex with 4-(4-fluorophenyl)-2-piperidin-1-yl-3-(1H-tetrazol-5-yl)-7,8-dihydro-5H-pyrano[4,3-b]pyridine, i.e. SMILES c1(ccc(cc1)F)c1c(c(N2CCCCC2)nc2c1COCC2)C1=NN=NN1 with IC50=0.42193 microM | | Descriptor: | (3M)-4-(4-fluorophenyl)-2-(piperidin-1-yl)-3-(1H-tetrazol-5-yl)-7,8-dihydro-5H-pyrano[4,3-b]pyridine, 1,2-ETHANEDIOL, FORMIC ACID, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Obst-Sander, U, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
4NA7
 
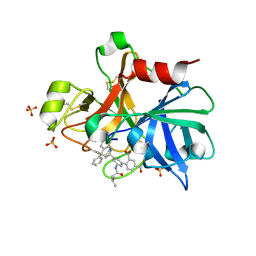 | | Factor XIA in complex with the inhibitor 3'-[(2S,4R)-6-carbamimidoyl-4-methyl-4-phenyl-1,2,3,4-tetrahydroquinolin-2-yl]-4-carbamoyl-5'-[(3-methylbutanoyl)amino]biphenyl-2-carboxylic acid | | Descriptor: | 3'-[(2S,4R)-6-carbamimidoyl-4-methyl-4-phenyl-1,2,3,4-tetrahydroquinolin-2-yl]-4-carbamoyl-5'-[(3-methylbutanoyl)amino]biphenyl-2-carboxylic acid, Coagulation factor XI, SULFATE ION | | Authors: | Wei, A. | | Deposit date: | 2013-10-21 | | Release date: | 2014-02-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Tetrahydroquinoline Derivatives as Potent and Selective Factor XIa Inhibitors.
J.Med.Chem., 57, 2014
|
|
7SCH
 
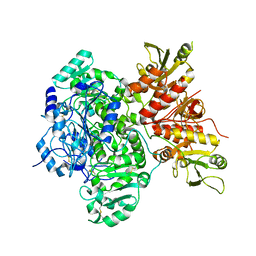 | | Cryo-EM structure of the human Exostosin-1 and Exostosin-2 heterodimer | | Descriptor: | Exostosin-1, Exostosin-2, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Li, H, Li, H. | | Deposit date: | 2021-09-28 | | Release date: | 2022-09-28 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for heparan sulfate co-polymerase action by the EXT1-2 complex.
Nat.Chem.Biol., 19, 2023
|
|
2QHN
 
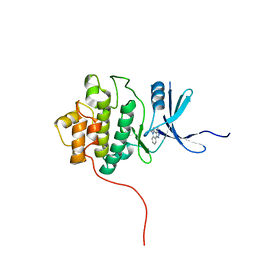 | | Crystal Structure of Chek1 in Complex with Inhibitor 1a | | Descriptor: | 5-ETHYL-3-METHYL-1,5-DIHYDRO-4H-PYRAZOLO[4,3-C]QUINOLIN-4-ONE, Serine/threonine-protein kinase Chk1 | | Authors: | Yan, Y, Munshi, S. | | Deposit date: | 2007-07-02 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Optimization of a pyrazoloquinolinone class of Chk1 kinase inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
9DG3
 
 | | ncPRC1RYBP Delta-linker mutant bound to singly modified H2AK119Ub nucleosome | | Descriptor: | DNA (187-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Godinez-Lopez, V, Valencia-Sanchez, M.I, Armache, J.P, Armache, K.-J. | | Deposit date: | 2024-09-01 | | Release date: | 2024-11-20 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Read-write mechanisms of H2A ubiquitination by Polycomb repressive complex 1.
Nature, 636, 2024
|
|
2YK7
 
 | | Structure of Neisseria LOS-specific sialyltransferase (NST), in complex with CMP-3F-Neu5Ac. | | Descriptor: | 1,2-ETHANEDIOL, CMP-N-ACETYLNEURAMINATE-BETA-GALACTOSAMIDE-ALPHA-2,3-SIALYLTRANSFERASE, CYTIDINE-5'-MONOPHOSPHATE-3-FLUORO-N-ACETYL-NEURAMINIC ACID, ... | | Authors: | Lin, L.Y.C, Rakic, B, Chiu, C.P.C, Lameignere, E, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2011-05-25 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure and Mechanism of the Lipooligosaccharide Sialyltransferase from Neisseria Meningitidis
J.Biol.Chem., 286, 2011
|
|
4N2R
 
 | | Crystal Structure of the alpha-L-arabinofuranosidase UmAbf62A from Ustilago maydis in complex with L-arabinofuranose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, alpha-L-arabinofuranose, ... | | Authors: | Siguier, B, Dumon, C, Mourey, L, Tranier, S. | | Deposit date: | 2013-10-06 | | Release date: | 2014-01-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | First Structural Insights into alpha-L-Arabinofuranosidases from the Two GH62 Glycoside Hydrolase Subfamilies.
J.Biol.Chem., 289, 2014
|
|
5LE5
 
 | | Native human 20S proteasome at 1.8 Angstrom | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Schrader, J, Henneberg, F, Mata, R, Tittmann, K, Schneider, T.R, Stark, H, Bourenkov, G, Chari, A. | | Deposit date: | 2016-06-29 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The inhibition mechanism of human 20S proteasomes enables next-generation inhibitor design.
Science, 353, 2016
|
|
5WPS
 
 | | Crystal structure HpiC1 Y101F | | Descriptor: | 1,2-ETHANEDIOL, 12-epi-hapalindole C/U synthase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.389 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
5LGQ
 
 | | Crystal structure of mouse CARM1 in complex with ligand P2C3s | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-propyl-oxolane-3,4-diol, 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, ... | | Authors: | Marechal, N, Troffer-Charlier, N, Cura, V, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-07-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Transition state mimics are valuable mechanistic probes for structural studies with the arginine methyltransferase CARM1.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4YL9
 
 | | Crystal Structure of wild-type of hsp14.1 from Sulfolobus solfatataricus P2 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Heat shock protein Hsp20 | | Authors: | Liu, L, Chen, J.Y, Yun, C.H. | | Deposit date: | 2015-03-05 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.353 Å) | | Cite: | Active-State Structures of a Small Heat-Shock Protein Revealed a Molecular Switch for Chaperone Function
Structure, 23, 2015
|
|
6RB6
 
 | | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-053 | | Descriptor: | 1,2-ETHANEDIOL, 3-[5-[(4~{a}~{R},8~{a}~{S})-3-cycloheptyl-4-oxidanylidene-4~{a},5,8,8~{a}-tetrahydrophthalazin-1-yl]-2-methoxy-phenyl]-~{N}-(furan-2-ylmethyl)prop-2-ynamide, FORMIC ACID, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2019-04-09 | | Release date: | 2019-07-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Alkynamide phthalazinones as a new class of TbrPDEB1 inhibitors (Part 2).
Bioorg.Med.Chem., 27, 2019
|
|
7AJH
 
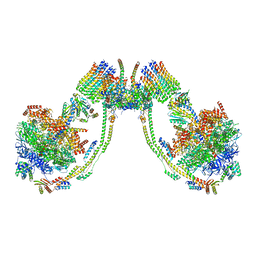 | | bovine ATP synthase dimer state3:state1 | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ATP synthase F(0) complex subunit B1, mitochondrial, ... | | Authors: | Spikes, T.E, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-09-29 | | Release date: | 2021-02-03 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (9.7 Å) | | Cite: | Interface mobility between monomers in dimeric bovine ATP synthase participates in the ultrastructure of inner mitochondrial membranes.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5T3P
 
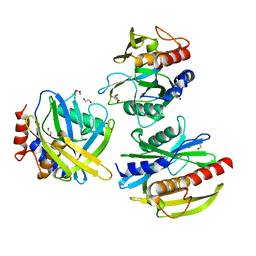 | | Crystal structure of Human Peroxisomal coenzyme A diphosphatase NUDT7 | | Descriptor: | 1,2-ETHANEDIOL, Peroxisomal coenzyme A diphosphatase NUDT7 | | Authors: | Srikannathasan, V, Nunez, C.A, Tallant, C, Siejka, P, Mathea, S, Kopec, J, Elkins, J.M, Burgess-Brown, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Huber, K. | | Deposit date: | 2016-08-26 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of Human Peroxisomal coenzyme A diphosphatase NUDT7
To Be Published
|
|
7YRK
 
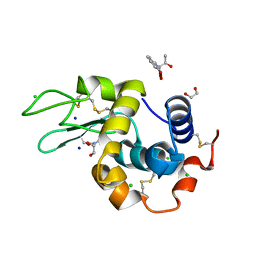 | | Crystal structure of the hen egg lysozyme-2-oxidobenzylidene-threoninato-copper (II) complex | | Descriptor: | (6~{S})-6-[(1~{R})-1-oxidanylethyl]-2,4-dioxa-7$l^{4}-aza-3$l^{3}-cupratricyclo[7.4.0.0^{3,7}]trideca-1(13),7,9,11-tetraen-5-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Unno, M, Furuya, T, Kitanishi, K, Akitsu, T. | | Deposit date: | 2022-08-10 | | Release date: | 2023-05-10 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | A novel hybrid protein composed of superoxide-dismutase-active Cu(II) complex and lysozyme.
Sci Rep, 13, 2023
|
|
2RA9
 
 | |
8R0B
 
 | | Cryo-EM structure of the cross-exon pre-B+ATP complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 5'ss oligo, NHP2-like protein 1, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-31 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
6FBP
 
 | | Human Methionine Adenosyltransferase II mutant (S114A) in P22121 crystal form | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Panmanee, J, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-12-19 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Control and regulation of S-Adenosylmethionine biosynthesis by the regulatory beta subunit and quinolone-based compounds.
Febs J., 286, 2019
|
|
5MU6
 
 | | Human N-myristoyltransferase (NMT1) with Myristoyl-CoA and IMP-1088 inhibitor bound | | Descriptor: | 1-[5-[3,4-bis(fluoranyl)-2-[2-(1,3,5-trimethylpyrazol-4-yl)ethoxy]phenyl]-1-methyl-indazol-3-yl]-~{N},~{N}-dimethyl-methanamine, GLYCEROL, Glycylpeptide N-tetradecanoyltransferase 1, ... | | Authors: | Perez-Dorado, I, Bell, A.S, Tate, E.W. | | Deposit date: | 2017-01-12 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Fragment-derived inhibitors of human N-myristoyltransferase block capsid assembly and replication of the common cold virus.
Nat Chem, 10, 2018
|
|
8R09
 
 | | Cryo-EM structure of the cross-exon pre-B+5'ss+ATPgammaS complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 5'ss oligo, NHP2-like protein 1, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-31 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
6R3C
 
 | |
