8AJ1
 
 | | SARS-CoV-2 Mpro in Complex with RK-107 | | Descriptor: | (2R,3S)-3-[[(2S)-3-cyclopropyl-2-[2-oxidanylidene-3-(phenylcarbamoylamino)pyridin-1-yl]propanoyl]amino]-N-methyl-2-oxidanyl-4-[(3S)-2-oxidanylidenepyrrolidin-3-yl]butanamide, 3C-like proteinase nsp5 | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2022-07-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | SARS-CoV-2 Mpro in Complex with RK-107
To Be Published
|
|
8TBL
 
 | | Tricomplex of RMC-7977, KRAS G12D, and CypA | | Descriptor: | (1R,5S,6r)-N-[(1P,7S,9S,13S,20M)-20-{5-(4-cyclopropylpiperazin-1-yl)-2-[(1S)-1-methoxyethyl]pyridin-3-yl}-21-ethyl-17,17-dimethyl-8,14-dioxo-15-oxa-4-thia-9,21,27,28-tetraazapentacyclo[17.5.2.1~2,5~.1~9,13~.0~22,26~]octacosa-1(24),2,5(28),19,22,25-hexaen-7-yl]-3-oxabicyclo[3.1.0]hexane-6-carboxamide, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Tomlinson, A.C.A, Chen, A, Knox, J.E, Yano, J.K. | | Deposit date: | 2023-06-28 | | Release date: | 2024-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Concurrent inhibition of oncogenic and wild-type RAS-GTP for cancer therapy.
Nature, 629, 2024
|
|
8TOR
 
 | | ACE2-peptide 2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Christie, M, Payne, R.J. | | Deposit date: | 2023-08-03 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of High Affinity Cyclic Peptide Ligands for Human ACE2 with SARS-CoV-2 Entry Inhibitory Activity.
Acs Chem.Biol., 19, 2024
|
|
7QF7
 
 | |
7A0P
 
 | | Neisseria gonorrhoeae Leucyl-tRNA Synthetase in Complex with Compound 11i | | Descriptor: | 1,2-ETHANEDIOL, Leucine--tRNA ligase, MAGNESIUM ION, ... | | Authors: | Pang, L, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Synthesis and structure-activity studies of novel anhydrohexitol-based Leucyl-tRNA synthetase inhibitors.
Eur.J.Med.Chem., 211, 2021
|
|
7LNR
 
 | | Structure of the avibactam-CDD-1 120 minute complex in imidazole and MPD | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-lactamase, ... | | Authors: | Smith, C.A, Vakulenko, S.B, Stewart, N.K. | | Deposit date: | 2021-02-08 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | In Crystallo Time-Resolved Interaction of the Clostridioides difficile CDD-1 enzyme with Avibactam Provides New Insights into the Catalytic Mechanism of Class D beta-lactamases.
Acs Infect Dis., 7, 2021
|
|
8BLR
 
 | | G13D mutant of KRAS4b (2-169) bound to GDP with the switch-I in fully open conformation | | Descriptor: | GTPase KRas, N-terminally processed, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Moche, M, Jungholm, O, Strandback, E, Ampah-Korsah, H, Nyman, T, Orwar, O. | | Deposit date: | 2022-11-10 | | Release date: | 2024-05-22 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Novel druggable space in human KRAS G13D discovered using structural bioinformatics and a P-loop targeting monoclonal antibody.
Sci Rep, 14, 2024
|
|
6IOT
 
 | | The ligand binding domain of Mlp24 with arginine | | Descriptor: | ARGININE, CALCIUM ION, Methyl-accepting chemotaxis protein | | Authors: | Takahashi, Y, Sumita, K, Nishiyama, S, Kawagishi, I, Imada, K. | | Deposit date: | 2018-10-31 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Calcium Ions Modulate Amino Acid Sensing of the Chemoreceptor Mlp24 ofVibrio cholerae.
J. Bacteriol., 201, 2019
|
|
8BPW
 
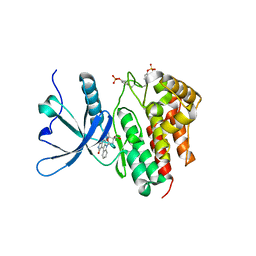 | | Crystal structure of JAK2 JH1 in complex with lestaurtinib | | Descriptor: | Lestaurtinib, Tyrosine-protein kinase JAK2 | | Authors: | Miao, Y, Haikarainen, T. | | Deposit date: | 2022-11-18 | | Release date: | 2023-11-29 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Functional and Structural Characterization of Clinical-Stage Janus Kinase 2 Inhibitors Identifies Determinants for Drug Selectivity.
J.Med.Chem., 67, 2024
|
|
7A1B
 
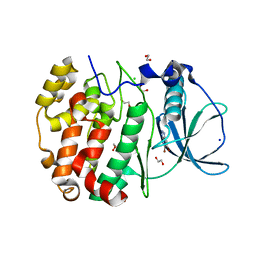 | | Crystal structure of human protein kinase CK2alpha' (CSNK2A2 gene product) in complex with the ATP-competitive inhibitor 5,6-dibromo-1H-triazolo[4,5-b]pyridine | | Descriptor: | 1,2-ETHANEDIOL, 5,6-dibromo-1H-triazolo[4,5-b]pyridine, CHLORIDE ION, ... | | Authors: | Niefind, K, Lindenblatt, D, Toelzer, C, Bretner, M, Chojnacki, K, Wielechowska, M, Winska, P. | | Deposit date: | 2020-08-12 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.287 Å) | | Cite: | Synthesis, biological properties and structural study of new halogenated azolo[4,5-b]pyridines as inhibitors of CK2 kinase.
Bioorg.Chem., 106, 2021
|
|
8TBM
 
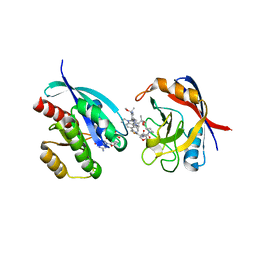 | | Tricomplex of RMC-7977, KRAS G12V, and CypA | | Descriptor: | (1R,5S,6r)-N-[(1P,7S,9S,13S,20M)-20-{5-(4-cyclopropylpiperazin-1-yl)-2-[(1S)-1-methoxyethyl]pyridin-3-yl}-21-ethyl-17,17-dimethyl-8,14-dioxo-15-oxa-4-thia-9,21,27,28-tetraazapentacyclo[17.5.2.1~2,5~.1~9,13~.0~22,26~]octacosa-1(24),2,5(28),19,22,25-hexaen-7-yl]-3-oxabicyclo[3.1.0]hexane-6-carboxamide, GLYCEROL, GTPase KRas, ... | | Authors: | Tomlinson, A.C.A, Chen, A, Knox, J.E, Yano, J.K. | | Deposit date: | 2023-06-28 | | Release date: | 2024-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Concurrent inhibition of oncogenic and wild-type RAS-GTP for cancer therapy.
Nature, 629, 2024
|
|
8TBN
 
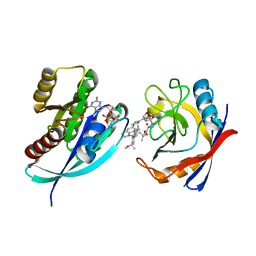 | | Tricomplex of RMC-7977, KRAS G12S, and CypA | | Descriptor: | (1R,5S,6r)-N-[(1P,7S,9S,13S,20M)-20-{5-(4-cyclopropylpiperazin-1-yl)-2-[(1S)-1-methoxyethyl]pyridin-3-yl}-21-ethyl-17,17-dimethyl-8,14-dioxo-15-oxa-4-thia-9,21,27,28-tetraazapentacyclo[17.5.2.1~2,5~.1~9,13~.0~22,26~]octacosa-1(24),2,5(28),19,22,25-hexaen-7-yl]-3-oxabicyclo[3.1.0]hexane-6-carboxamide, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Tomlinson, A.C.A, Chen, A, Knox, J.E, Yano, J.K. | | Deposit date: | 2023-06-28 | | Release date: | 2024-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Concurrent inhibition of oncogenic and wild-type RAS-GTP for cancer therapy.
Nature, 629, 2024
|
|
6IP7
 
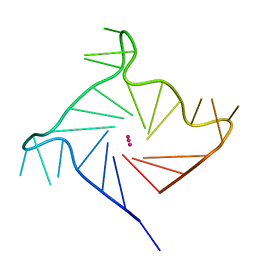 | | Structure of human telomeric DNA with 5-Selenophene-modified deoxyuridine at residue 11 | | Descriptor: | DNA (22-MER), POTASSIUM ION | | Authors: | Saikrishnan, K, Nuthanakanti, A, Srivatsan, S.G, Ahmad, I. | | Deposit date: | 2018-11-02 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Probing G-quadruplex topologies and recognition concurrently in real time and 3D using a dual-app nucleoside probe.
Nucleic Acids Res., 47, 2019
|
|
7LOL
 
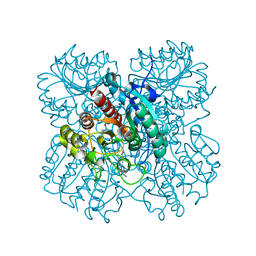 | | The structure of Agmatinase from E. Coli at 1.8 A displaying urea and agmatine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AGMATINE, Agmatinase, ... | | Authors: | Maturana, P, Figueroa, M, Gonzalez-Ordenes, F, Villalobos, P, Martinez-Oyanedel, J, Uribe, E.A, Castro-Fernandez, V. | | Deposit date: | 2021-02-10 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Escherichia coli Agmatinase: Catalytic Mechanism and Residues Relevant for Substrate Specificity.
Int J Mol Sci, 22, 2021
|
|
7LOX
 
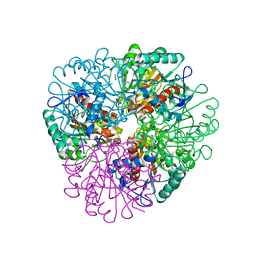 | | The structure of Agmatinase from E. Coli at 3.2 A displaying guanidine in the active site | | Descriptor: | Agmatinase, GUANIDINE, MANGANESE (II) ION | | Authors: | Maturana, P, Figueroa, M, Gonzalez-Ordenes, F, Villalobos, P, Martinez-Oyanedel, J, Uribe, E.A, Castro-Fernandez, V. | | Deposit date: | 2021-02-11 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of Escherichia coli Agmatinase: Catalytic Mechanism and Residues Relevant for Substrate Specificity.
Int J Mol Sci, 22, 2021
|
|
7QFA
 
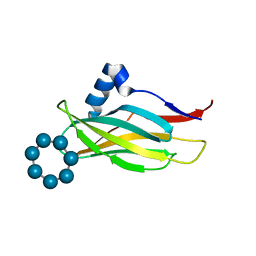 | |
7LUO
 
 | | N-terminus of Skp2 bound to Cyclin A | | Descriptor: | S-phase kinase-associated protein 2,Cyclin-A2, Skp2 Motif 1 uncharacterized fragment 1, Skp2 Motif 1 uncharacterized fragment 2 | | Authors: | Kelso, S, Ceccarelli, D.F, Sicheri, F. | | Deposit date: | 2021-02-22 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Bipartite binding of the N terminus of Skp2 to cyclin A.
Structure, 29, 2021
|
|
6C64
 
 | | Crystal Structure of the Mango-II Fluorescent Aptamer Bound to TO3-Biotin | | Descriptor: | 1-methyl-4-[(1E)-3-(3-methyl-1,3-benzothiazol-3-ium-2-yl)prop-1-en-1-yl]quinolin-1-ium, POTASSIUM ION, RNA (32-MER), ... | | Authors: | Trachman, R.J, Ferre-D'Amare, A.R. | | Deposit date: | 2018-01-17 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.00014877 Å) | | Cite: | Crystal Structures of the Mango-II RNA Aptamer Reveal Heterogeneous Fluorophore Binding and Guide Engineering of Variants with Improved Selectivity and Brightness.
Biochemistry, 57, 2018
|
|
6IR1
 
 | | Crystal structure of red fluorescent protein mCherry complexed with the nanobody LaM4 at 1.9 Angstron resolution | | Descriptor: | MCherry fluorescent protein, mCherry's nanobody LaM4 | | Authors: | Ding, Y, Wang, Z.Y, Hu, R.T, Chen, X. | | Deposit date: | 2018-11-09 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.919 Å) | | Cite: | Structural insights into the binding of nanobodies LaM2 and LaM4 to the red fluorescent protein mCherry.
Protein Sci., 30, 2021
|
|
6ZLR
 
 | |
8TCA
 
 | | Structure of a mouse IgG antibody antigen-binding fragment (Fab) targeting N6-methyladenosine (m6A), an RNA modification, no ligand | | Descriptor: | 1,2-ETHANEDIOL, m6A binding IgG Fab, heavy chain, ... | | Authors: | Angelo, M.C, Aoki, S.T. | | Deposit date: | 2023-06-30 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure of a mouse IgG antibody antigen-binding fragment (Fab) targeting N6-methyladenosine (m6A), an RNA modification
To Be Published
|
|
7LRQ
 
 | | Crystal structure of human SFPQ/NONO heterodimer, conserved DBHS region | | Descriptor: | CHLORIDE ION, Non-POU domain-containing octamer-binding protein, Splicing factor, ... | | Authors: | Marshall, A.C, Bond, C.S, Mohnen, I. | | Deposit date: | 2021-02-17 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Paraspeckle subnuclear bodies depend on dynamic heterodimerisation of DBHS RNA-binding proteins via their structured domains.
J.Biol.Chem., 298, 2022
|
|
8TOT
 
 | | ACE2-peptide2 complex crystal form 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Christie, M, Payne, R.J. | | Deposit date: | 2023-08-03 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of High Affinity Cyclic Peptide Ligands for Human ACE2 with SARS-CoV-2 Entry Inhibitory Activity.
Acs Chem.Biol., 19, 2024
|
|
7LRU
 
 | | Crystal structure of SFPQ-NONO-SFPQ chimeric protein homodimer | | Descriptor: | Splicing factor, proline- and glutamine-rich,Isoform 2 of Non-POU domain-containing octamer-binding protein,Isoform Short of Splicing factor, proline- and glutamine-rich | | Authors: | Marshall, A.C, Bond, C.S, Mohnen, I, Knott, G.J. | | Deposit date: | 2021-02-17 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of SFPQ-NONO-SFPQ chimeric protein homodimer
To Be Published
|
|
6IRW
 
 | | Crystal structure of the human cap-specific adenosine methyltransferase bound to SAH | | Descriptor: | Phosphorylated CTD-interacting factor 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-14 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase.
Science, 363, 2019
|
|
