6OZM
 
 | |
6OZO
 
 | |
6P28
 
 | |
6P25
 
 | | Structure of S. cerevisiae protein O-mannosyltransferase Pmt1-Pmt2 complex bound to the sugar donor and a peptide acceptor | | Descriptor: | (3R)-3,31-dimethyl-7,11,15,19,23,27-hexamethylidenedotriacont-31-en-1-yl dihydrogen phosphate, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bai, L, Li, H. | | Deposit date: | 2019-05-21 | | Release date: | 2019-07-10 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the eukaryotic protein O-mannosyltransferase Pmt1-Pmt2 complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6OZ5
 
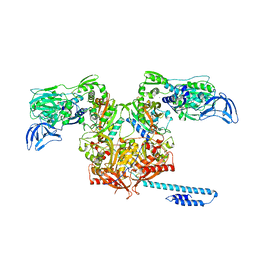 | | Escherichia coli tRNA synthetase in complex with compound 3 | | Descriptor: | 1,2-ETHANEDIOL, 2-({[(2S)-1-cyclohexylpropan-2-yl]amino}methyl)phenol, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kahne, D, Baidin, V, Owens, T.W. | | Deposit date: | 2019-05-15 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Simple Secondary Amines Inhibit Growth of Gram-Negative Bacteria through Highly Selective Binding to Phenylalanyl-tRNA Synthetase.
J.Am.Chem.Soc., 143, 2021
|
|
6OZG
 
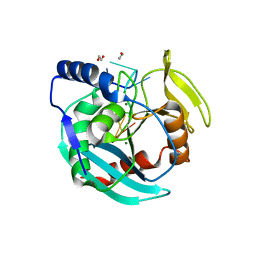 | |
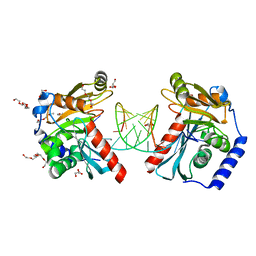
6OZN
 
 | |
5DA9
 
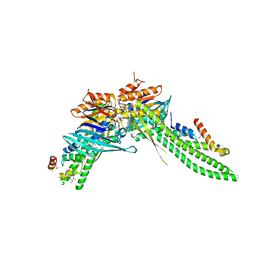 | | ATP-gamma-S bound Rad50 from Chaetomium thermophilum in complex with the Rad50-binding domain of Mre11 | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Putative double-strand break protein, ... | | Authors: | Seifert, F.U, Lammens, K, Stoehr, G, Kessler, B, Hopfner, K.-P. | | Deposit date: | 2015-08-19 | | Release date: | 2016-03-02 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural mechanism of ATP-dependent DNA binding and DNA end bridging by eukaryotic Rad50.
Embo J., 35, 2016
|
|
5DAC
 
 | | ATP-gamma-S bound Rad50 from Chaetomium thermophilum in complex with DNA | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, DNA (5'-D(P*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*C)-3'), DNA (5'-D(P*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*G)-3'), ... | | Authors: | Seifert, F.U, Lammens, K, Stoehr, G, Kessler, B, Hopfner, K.-P. | | Deposit date: | 2015-08-19 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Structural mechanism of ATP-dependent DNA binding and DNA end bridging by eukaryotic Rad50.
Embo J., 35, 2016
|
|
7P4L
 
 | | Crystal structure of the trimeric ectodomain of archaeal Fusexin1 (Fsx1) | | Descriptor: | CALCIUM ION, CHLORIDE ION, Fusexin1, ... | | Authors: | Nishio, S, Tunyasuvunakool, K, Jumper, J, De Sanctis, D, Jovine, L. | | Deposit date: | 2021-07-12 | | Release date: | 2022-07-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of archaeal fusexins homologous to eukaryotic HAP2/GCS1 gamete fusion proteins.
Nat Commun, 13, 2022
|
|
7POJ
 
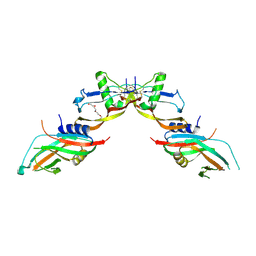 | | Prodomain bound BMP10 crystal form 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bone morphogenetic protein 10, TETRAETHYLENE GLYCOL | | Authors: | Guo, J, Yu, M, Li, W. | | Deposit date: | 2021-09-09 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of BMPRII extracellular domain in binary and ternary receptor complexes with BMP10.
Nat Commun, 13, 2022
|
|
7POI
 
 | | Prodomain bound BMP10 crystal form 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bone morphogenetic protein 10, D(-)-TARTARIC ACID | | Authors: | Guo, J, Yu, M, Li, W. | | Deposit date: | 2021-09-09 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of BMPRII extracellular domain in binary and ternary receptor complexes with BMP10.
Nat Commun, 13, 2022
|
|
7PPB
 
 | |
6IXT
 
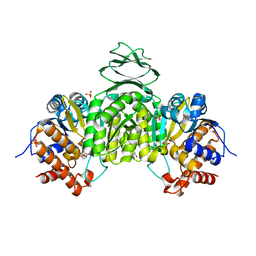 | | Crystal structure of isocitrate dehydrogenase from Ostreococcus tauri in complex with NAD+ and Mg2+ | | Descriptor: | GLYCEROL, Isocitrate dehydrogenase, MAGNESIUM ION, ... | | Authors: | Zhu, G.P, Tang, W.G, Wang, P. | | Deposit date: | 2018-12-12 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structures of NAD + -linked isocitrate dehydrogenase from the green alga Ostreococcus tauri and its evolutionary relationship with eukaryotic NADP + -linked homologs.
Arch.Biochem.Biophys., 708, 2021
|
|
3FAP
 
 | |
5XAJ
 
 | |
1ZYZ
 
 | | Structures of Yeast Ribonucloetide Reductase I | | Descriptor: | GLYCINE, Ribonucleoside-diphosphate reductase large chain 1 | | Authors: | Xu, H, Faber, C, Uchiki, T, Fairman, J.W, Racca, J, Dealwis, C. | | Deposit date: | 2005-06-13 | | Release date: | 2006-03-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of eukaryotic ribonucleotide reductase I provide insights into dNTP regulation
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
6F6D
 
 | | The catalytic domain of KDM6B in complex with H3(17-33)K18IA21M peptide | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, Histone 3 peptide H3(17-33)K18IA21M, ... | | Authors: | Jones, S.E, Olsen, L, Gajhede, M. | | Deposit date: | 2017-12-05 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.81810677 Å) | | Cite: | Structural Basis of Histone Demethylase KDM6B Histone 3 Lysine 27 Specificity.
Biochemistry, 57, 2018
|
|
4W5I
 
 | | Crystal structure of human tankyrase 2 in complex with 1-methyl-7-phenyl-1,2,3,4,5,6-hexahydro-1,6- naphthyridin-5-one | | Descriptor: | 1-methyl-7-phenyl-2,3,4,6-tetrahydro-1,6-naphthyridin-5(1H)-one, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Lehtio, L. | | Deposit date: | 2014-08-18 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-based design, synthesis and evaluation in vitro of arylnaphthyridinones, arylpyridopyrimidinones and their tetrahydro derivatives as inhibitors of the tankyrases.
Bioorg.Med.Chem., 23, 2015
|
|
6FIL
 
 | | DDR1, 2-[8-(1H-indazole-5-carbonyl)-4-oxo-1-phenyl-1,3,8-triazaspiro[4.5]decan-3-yl]-N-methylacetamide, 1.730A, P212121, Rfree=24.5% | | Descriptor: | 2-[8-(2~{H}-indazol-5-ylcarbonyl)-4-oxidanylidene-1-phenyl-1,3,8-triazaspiro[4.5]decan-3-yl]-~{N}-methyl-ethanamide, Epithelial discoidin domain-containing receptor 1, IODIDE ION | | Authors: | Stihle, M, Richter, H, Benz, J, Kuhn, B, Rudolph, M.G. | | Deposit date: | 2018-01-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | DNA-Encoded Library-Derived DDR1 Inhibitor Prevents Fibrosis and Renal Function Loss in a Genetic Mouse Model of Alport Syndrome.
Acs Chem.Biol., 14, 2019
|
|
4USZ
 
 | | Crystal structure of the first bacterial vanadium dependant iodoperoxidase | | Descriptor: | SODIUM ION, VANADATE ION, VANADIUM-DEPENDENT HALOPEROXIDASE | | Authors: | Rebuffet, E, Delage, L, Fournier, J.B, Rzonca, J, Potin, P, Michel, G, Czjzek, M, Leblanc, C. | | Deposit date: | 2014-07-17 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Bacterial Vanadium Iodoperoxidase from the Marine Flavobacteriaceae Zobellia Galactanivorans Reveals Novel Molecular and Evolutionary Features of Halide Specificity in This Enzyme Family.
Appl.Environ.Microbiol., 80, 2014
|
|
6FEX
 
 | | DDR1, 2-[4-bromo-2-oxo-1'-(1H-pyrazolo[4,3-b]pyridine-5-carbonyl)spiro[indole-3,4'-piperidine]-1-yl]-N-(2,2,2-trifluoroethyl)acetamide, 1.291A, P212121, Rfree=17.4% | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[4-bromanyl-2-oxidanylidene-1'-(1~{H}-pyrazolo[4,3-b]pyridin-5-ylcarbonyl)spiro[indole-3,4'-piperidine]-1-yl]-~{N}-[2,2,2-tris(fluoranyl)ethyl]ethanamide, CHLORIDE ION, ... | | Authors: | Stihle, M, Richter, H, Benz, J, Kuhn, B, Rudolph, M.G. | | Deposit date: | 2018-01-03 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.291 Å) | | Cite: | DNA-Encoded Library-Derived DDR1 Inhibitor Prevents Fibrosis and Renal Function Loss in a Genetic Mouse Model of Alport Syndrome.
Acs Chem.Biol., 14, 2019
|
|
1ZZD
 
 | | Structures of Yeast Ribonucleotide Reductase I | | Descriptor: | Ribonucleoside-diphosphate reductase large chain 1, Ribonucleoside-diphosphate reductase small chain 2 | | Authors: | Xu, H, Faber, C, Uchiki, T, Fairman, J.W, Racca, J, Dealwis, C. | | Deposit date: | 2005-06-13 | | Release date: | 2006-03-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of eukaryotic ribonucleotide reductase I provide insights into dNTP regulation
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
6FIN
 
 | | DDR1, 3-[(3-cyclopropyl-1,2,4-oxadiazol-5-yl)methyl]-8-(1H-indazole-5-carbonyl)-1-phenyl-1,3,8-triazaspiro[4.5]decan-4-one, 1.670A, P1211, Rfree=22.8% | | Descriptor: | 3-[(3-cyclopropyl-1,2,4-oxadiazol-5-yl)methyl]-8-(1~{H}-indazol-5-ylcarbonyl)-1-phenyl-1,3,8-triazaspiro[4.5]decan-4-one, Epithelial discoidin domain-containing receptor 1, IODIDE ION | | Authors: | Stihle, M, Richter, H, Benz, J, Kuhn, B, Rudolph, M.G. | | Deposit date: | 2018-01-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | DNA-Encoded Library-Derived DDR1 Inhibitor Prevents Fibrosis and Renal Function Loss in a Genetic Mouse Model of Alport Syndrome.
Acs Chem.Biol., 14, 2019
|
|
6FIQ
 
 | | DDR1, 1-(1H-indazole-5-carbonyl)-5'-methoxy-1'-[2-oxo-2-[(2S)-2-(trifluoromethyl)pyrrolidin-1-yl]ethyl]spiro[piperidine-4,3'-pyrrolo[3,2-b]pyridine]-2'-one, 1.790A, P212121, Rfree=23.8% | | Descriptor: | 1-(1~{H}-indazol-5-ylcarbonyl)-5'-methoxy-1'-[2-oxidanylidene-2-[(2~{S})-2-(trifluoromethyl)pyrrolidin-1-yl]ethyl]spiro[piperidine-4,3'-pyrrolo[3,2-b]pyridine]-2'-one, CHLORIDE ION, Epithelial discoidin domain-containing receptor 1 | | Authors: | Stihle, M, Richter, H, Benz, J, Kuhn, B, Rudolph, M.G. | | Deposit date: | 2018-01-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | DNA-Encoded Library-Derived DDR1 Inhibitor Prevents Fibrosis and Renal Function Loss in a Genetic Mouse Model of Alport Syndrome.
Acs Chem.Biol., 14, 2019
|
|
