3VS0
 
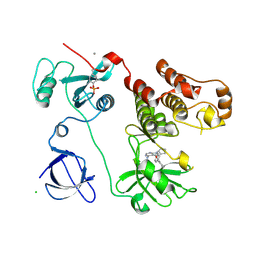 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor N-[4-(4-amino-7-cyclopentyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)phenyl]benzamide | | Descriptor: | CALCIUM ION, CHLORIDE ION, N-[4-(4-amino-7-cyclopentyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)phenyl]benzamide, ... | | Authors: | Kuratani, M, Honda, K, Tomabechi, Y, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.934 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
3VS6
 
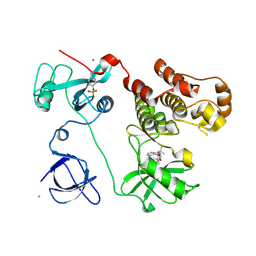 | | Crystal structure of HCK complexed with a pyrazolo-pyrimidine inhibitor tert-butyl {4-[4-amino-1-(propan-2-yl)-1H-pyrazolo[3,4-d]pyrimidin-3-yl]-2-methoxyphenyl}carbamate | | Descriptor: | CALCIUM ION, CHLORIDE ION, Tyrosine-protein kinase HCK, ... | | Authors: | Kuratani, M, Honda, K, Tomabechi, Y, Toyama, M, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.373 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
3VS3
 
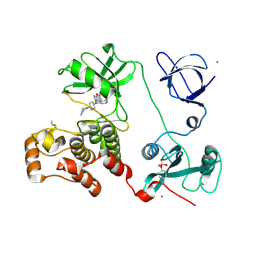 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine | | Descriptor: | 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kuratani, M, Tomaebchi, Y, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
3VS5
 
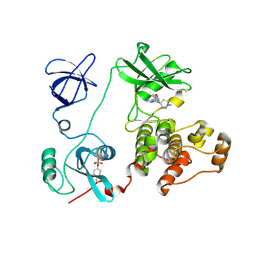 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor 7-(1-methylpiperidin-4-yl)-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine | | Descriptor: | 7-(1-methylpiperidin-4-yl)-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, CALCIUM ION, Tyrosine-protein kinase HCK | | Authors: | Kuratani, M, Tomabechi, Y, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
4EIK
 
 | |
2LQW
 
 | |
2LQN
 
 | | Solution structure of CRKL | | Descriptor: | Crk-like protein | | Authors: | Jankowski, W, Saleh, T, Kalodimos, C. | | Deposit date: | 2012-03-09 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Domain organization differences explain Bcr-Abl's preference for CrkL over CrkII.
Nat.Chem.Biol., 8, 2012
|
|
2LP5
 
 | | Native Structure of the Fyn SH3 A39V/N53P/V55L | | Descriptor: | Tyrosine-protein kinase Fyn | | Authors: | Neudecker, P, Robustelli, P, Cavalli, A, Vendruscolo, M, Kay, L.E. | | Deposit date: | 2012-02-06 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of an intermediate state in protein folding and aggregation.
Science, 336, 2012
|
|
4D8D
 
 | |
4D8K
 
 | |
2LMJ
 
 | | Itk-sh3 | | Descriptor: | Tyrosine-protein kinase ITK/TSK | | Authors: | Kristiansen, P, Bie Andersen, T, Huszenicza, Z, Andreotti, A.H, Spurkland, A. | | Deposit date: | 2011-12-05 | | Release date: | 2012-12-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The SH3 domains of the Tec family kinase Itk and the Src family kinase Lck compete for adjacent sites on T-cell specific adapter protein
To be Published
|
|
3UF4
 
 | |
4A63
 
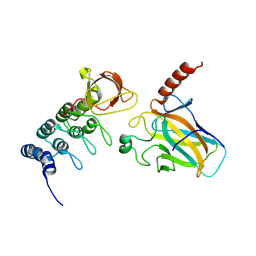 | | Crystal structure of the p73-ASPP2 complex at 2.6A resolution | | Descriptor: | ACETATE ION, APOPTOSIS STIMULATING OF P53 PROTEIN 2, TUMOUR PROTEIN 73, ... | | Authors: | Canning, P, Sharpe, T, Krojer, T, Savitsky, P, Cooper, C.D.O, Salah, E, Keates, T, Muniz, J, Vollmar, M, von Delft, F, Weigelt, J, Arrowsmith, C, Bountra, C, Edwards, A, Bullock, A.N. | | Deposit date: | 2011-10-31 | | Release date: | 2011-12-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural Basis for Aspp2 Recognition by the Tumor Suppressor P73.
J.Mol.Biol., 423, 2012
|
|
3UAT
 
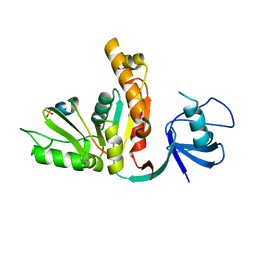 | |
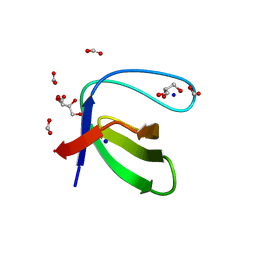
3UA6
 
 | |
3UA7
 
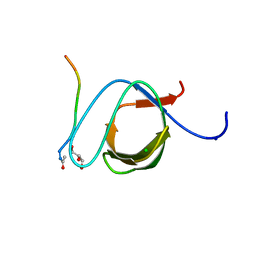 | | Crystal Structure of the Human Fyn SH3 domain in complex with a peptide from the Hepatitis C virus NS5A-protein | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Martin-Garcia, J.M, Ruiz-Sanz, J, Luque, I, Camara-Artigas, A. | | Deposit date: | 2011-10-21 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The promiscuous binding of the Fyn SH3 domain to a peptide from the NS5A protein.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3TVT
 
 | |
2LJ3
 
 | |
3THK
 
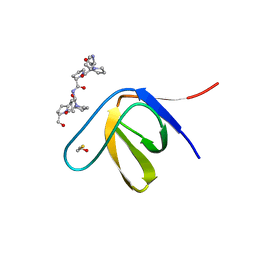 | | Structure of SH3 chimera with a type II ligand linked to the chain C-terminal | | Descriptor: | BETA-MERCAPTOETHANOL, Proline-rich peptide, SULFATE ION, ... | | Authors: | Gabdulkhakov, A.G, Gushchina, L.V, Nikulin, A.D, Nikonov, S.V, Filimonov, V.V. | | Deposit date: | 2011-08-19 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-Resolution Crystal Structure of Spectrin SH3 Domain Fused with a Proline-Rich Peptide.
J.Biomol.Struct.Dyn., 29, 2011
|
|
2LCS
 
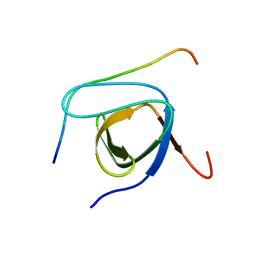 | | Yeast Nbp2p SH3 domain in complex with a peptide from Ste20p | | Descriptor: | NAP1-binding protein 2, Serine/threonine-protein kinase STE20 | | Authors: | Gorelik, M, Davidson, A.R. | | Deposit date: | 2011-05-07 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Distinct Peptide Binding Specificities of Src Homology 3 (SH3) Protein Domains Can Be Determined by Modulation of Local Energetics across the Binding Interface.
J.Biol.Chem., 287, 2012
|
|
3REB
 
 | |
3REA
 
 | |
3RBB
 
 | | HIV-1 NEF protein in complex with engineered HCK SH3 domain | | Descriptor: | 1,2-ETHANEDIOL, Protein Nef, Tyrosine-protein kinase HCK | | Authors: | Horenkamp, F.A, Schulte, A, Weyand, M, Geyer, M. | | Deposit date: | 2011-03-29 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Conformation of the Dileucine-Based Sorting Motif in HIV-1 Nef Revealed by Intermolecular Domain Assembly.
Traffic, 12, 2011
|
|
3QWX
 
 | | CED-2 1-174 | | Descriptor: | Cell death abnormality protein 2, SULFATE ION | | Authors: | Kang, Y, Sun, J, Liu, Y. | | Deposit date: | 2011-02-28 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of the cell corpse engulfment protein CED-2 in Caenorhabditis elegans.
Biochem.Biophys.Res.Commun., 410, 2011
|
|
3QWY
 
 | | CED-2 | | Descriptor: | Cell death abnormality protein 2, GLYCEROL, SULFATE ION | | Authors: | Kang, Y, Sun, J, Liu, Y, Sun, D, Hu, Y, Liu, Y.F. | | Deposit date: | 2011-02-28 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structure of the cell corpse engulfment protein CED-2 in Caenorhabditis elegans.
Biochem.Biophys.Res.Commun., 410, 2011
|
|
