6VU3
 
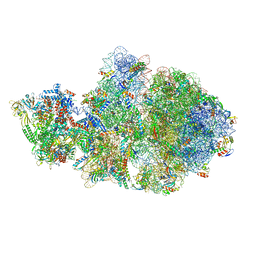 | | Cryo-EM structure of Escherichia coli transcription-translation complex A (TTC-A) containing mRNA with a 12 nt long spacer | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R. | | Deposit date: | 2020-02-14 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6VZJ
 
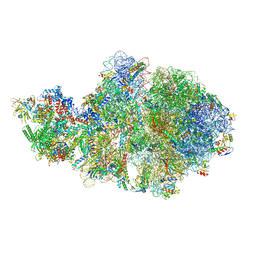 | | Escherichia coli transcription-translation complex A1 (TTC-A1) containing mRNA with a 15 nt long spacer, fMet-tRNAs at E-site and P-site, and lacking transcription factor NusG | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | Deposit date: | 2020-02-28 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6VYZ
 
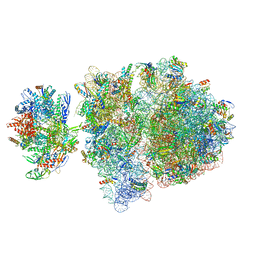 | | Escherichia coli transcription-translation complex C6 (TTC-C6) containing mRNA with a 21 nt long spacer, NusA, and fMet-tRNAs at E-site and P-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | Deposit date: | 2020-02-27 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (9.9 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6VYR
 
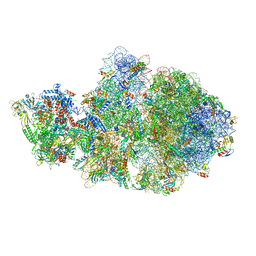 | | Escherichia coli transcription-translation complex A1 (TTC-A1) containing an 18 nt long mRNA spacer, NusG, and fMet-tRNAs at E-site and P-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | Deposit date: | 2020-02-27 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
8CEE
 
 | | Rnase R bound to a 30S degradation intermediate (State I - head-turning) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Paternoga, H, Dimitrova-Paternoga, L, Wilson, D.N. | | Deposit date: | 2023-02-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of ribosomal 30S subunit degradation by RNase R.
Nature, 626, 2024
|
|
8CED
 
 | | Rnase R bound to a 30S degradation intermediate (State I - head-turning) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Paternoga, H, Dimitrova-Paternoga, L, Wilson, D.N. | | Deposit date: | 2023-02-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Structural basis of ribosomal 30S subunit degradation by RNase R.
Nature, 626, 2024
|
|
8CDU
 
 | | Rnase R bound to a 30S degradation intermediate (main state) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Paternoga, H, Dimitrova-Paternoga, L, Wilson, D.N. | | Deposit date: | 2023-02-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of ribosomal 30S subunit degradation by RNase R.
Nature, 626, 2024
|
|
8CEC
 
 | | Rnase R bound to a 30S degradation intermediate (State I - head-turning) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Paternoga, H, Dimitrova-Paternoga, L, Wilson, D.N. | | Deposit date: | 2023-02-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structural basis of ribosomal 30S subunit degradation by RNase R.
Nature, 626, 2024
|
|
6VYT
 
 | | Escherichia coli transcription-translation complex A2 (TTC-A2) containing a 15 nt long mRNA spacer, NusG, and fMet-tRNAs at P-site and E-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Ebright, R.H, Su, M. | | Deposit date: | 2020-02-27 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
8CDV
 
 | | Rnase R bound to a 30S degradation intermediate (state II) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Paternoga, H, Dimitrova-Paternoga, L, Wilson, D.N. | | Deposit date: | 2023-02-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.73 Å) | | Cite: | Structural basis of ribosomal 30S subunit degradation by RNase R.
Nature, 626, 2024
|
|
6VZ2
 
 | | Escherichia coli transcription-translation complex D1 (TTC-D1) containing mRNA with a 27 nt long spacer, NusG, and fMet-tRNAs at E-site and P-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | Deposit date: | 2020-02-27 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
8CAS
 
 | | Cryo-EM structure of native Otu2-bound ubiquitinated 48S initiation complex (partial) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Ikeuchi, K, Buschauer, R, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2023-01-24 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for recognition and deubiquitination of 40S ribosomes by Otu2.
Nat Commun, 14, 2023
|
|
7SYS
 
 | | Structure of the delta dII IRES eIF2-containing 48S initiation complex, closed conformation. Structure 12(delta dII). | | Descriptor: | 18S rRNA, Eukaryotic translation initiation factor 1A, X-chromosomal, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-27 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYR
 
 | | Structure of the wt IRES eIF2-containing 48S initiation complex, closed conformation. Structure 12(wt). | | Descriptor: | 18S rRNA, Eukaryotic translation initiation factor 1A, X-chromosomal, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-27 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
2R93
 
 | | Elongation complex of RNA polymerase II with a hepatitis delta virus-derived RNA stem loop | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Lehmann, E, Brueckner, F, Cramer, P. | | Deposit date: | 2007-09-12 | | Release date: | 2007-11-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Molecular basis of RNA-dependent RNA polymerase II activity.
Nature, 450, 2007
|
|
8CRO
 
 | | Cryo-EM structure of Pyrococcus furiosus transcription elongation complex | | Descriptor: | DNA Non-Template Strand, DNA Template Strand, DNA-directed RNA polymerase subunit Rpo10, ... | | Authors: | Tarau, D.M, Grunberger, F, Reichelt, R, Heiss, F.B, Pilsl, M, Hausner, W, Engel, C, Grohmann, D. | | Deposit date: | 2023-03-08 | | Release date: | 2024-04-17 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of archaeal RNA polymerase transcription elongation and Spt4/5 recruitment.
Nucleic Acids Res., 52, 2024
|
|
7UBN
 
 | |
7UND
 
 | | Pol II-DSIF-SPT6-PAF1c-TFIIS-nucleosome complex (stalled at +38) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB3, DNA-directed RNA polymerase II subunit RPB7, ... | | Authors: | Filipovski, M, Vos, S.M, Farnung, L. | | Deposit date: | 2022-04-10 | | Release date: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of nucleosome retention during transcription elongation.
Science, 376, 2022
|
|
7UNC
 
 | | Pol II-DSIF-SPT6-PAF1c-TFIIS complex with rewrapped nucleosome | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB3, DNA-directed RNA polymerase II subunit RPB7, ... | | Authors: | Filipovski, M, Vos, S.M, Farnung, L. | | Deposit date: | 2022-04-10 | | Release date: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of nucleosome retention during transcription elongation.
Science, 376, 2022
|
|
7UIF
 
 | | Mediator-PIC Early (Core B) | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Gorbea Colon, J.J, Chen, S.-F, Tsai, K.L, Murakami, K. | | Deposit date: | 2022-03-29 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis of a transcription pre-initiation complex on a divergent promoter.
Mol.Cell, 83, 2023
|
|
7UIO
 
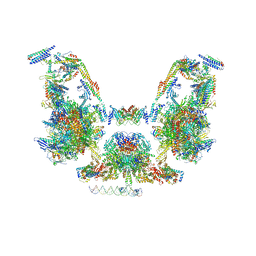 | | Mediator-PIC Early (Composite Model) | | Descriptor: | ALANINE, ASPARTIC ACID, DNA (37-MER), ... | | Authors: | Gorbea Colon, J.J, Chen, S.-F, Tsai, K.L, Murakami, K. | | Deposit date: | 2022-03-29 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of a transcription pre-initiation complex on a divergent promoter.
Mol.Cell, 83, 2023
|
|
7UI9
 
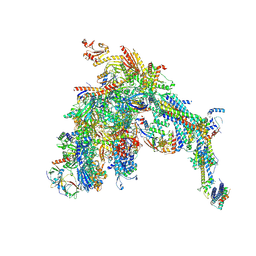 | | Core Mediator-PICearly (Copy A) | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Gorbea Colon, J.J, Chen, S.-F, Tsai, K.L, Murakami, K. | | Deposit date: | 2022-03-28 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of a transcription pre-initiation complex on a divergent promoter.
Mol.Cell, 83, 2023
|
|
2WAQ
 
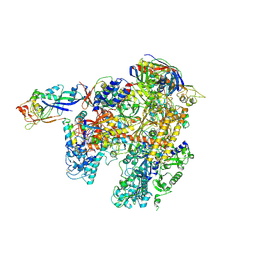 | | The complete structure of the archaeal 13-subunit DNA-directed RNA Polymerase | | Descriptor: | DNA-DIRECTED RNA POLYMERASE RPO10 SUBUNIT, DNA-DIRECTED RNA POLYMERASE RPO11 SUBUNIT, DNA-DIRECTED RNA POLYMERASE RPO12 SUBUNIT, ... | | Authors: | Korkhin, Y, Unligil, U.M, Littlefield, O, Nelson, P.J, Stuart, D.I, Sigler, P.B, Bell, S.D, Abrescia, N.G.A. | | Deposit date: | 2009-02-11 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Evolution of complex RNA polymerases: the complete archaeal RNA polymerase structure.
Plos Biol., 7, 2009
|
|
2VUM
 
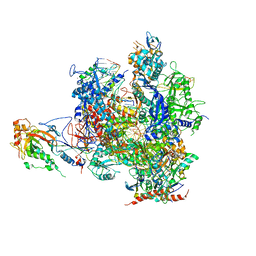 | | Alpha-amanitin inhibited complete RNA polymerase II elongation complex | | Descriptor: | 5'-D(*AP*AP*AP*CP*TP*AP*CP*TP*TP*GP *AP*GP*CP*T)-3', 5'-D(*AP*GP*CP*TP*CP*AP*AP*GP*TP*AP *GP*TP*TP*AP*CP*GP*CP*CP*BRUP*GP*GP*TP*CP*AP*TP*T)-3', 5'-R(*AP*AP*AP*GP*AP*CP*CP*AP*GP*GP*C)-3', ... | | Authors: | Brueckner, F, Cramer, P. | | Deposit date: | 2008-05-27 | | Release date: | 2008-06-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis of Transcription Inhibition by Alpha-Amanitin and Implications for RNA Polymerase II Translocation.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2WB1
 
 | | The complete structure of the archaeal 13-subunit DNA-directed RNA Polymerase | | Descriptor: | DNA-DIRECTED RNA POLYMERASE RPO10 SUBUNIT, DNA-DIRECTED RNA POLYMERASE RPO11 SUBUNIT, DNA-DIRECTED RNA POLYMERASE RPO12 SUBUNIT, ... | | Authors: | Korkhin, Y, Unligil, U.M, Littlefield, O, Nelson, P.J, Stuart, D.I, Sigler, P.B, Bell, S.D, Abrescia, N.G.A. | | Deposit date: | 2009-02-19 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Evolution of Complex RNA Polymerase: The Complete Archaeal RNA Polymerase Structure
Plos Biol., 7, 2009
|
|
