6CN1
 
 | | 2.75 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Pseudomonas putida in Complex with Uridine-diphosphate-2(n-acetylglucosaminyl) butyric acid, (2R)-2-(phosphonooxy)propanoic acid and Magnesium | | Descriptor: | (2R)-2-(phosphonooxy)propanoic acid, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Cardona-Correa, A, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-03-06 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | 2.75 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Pseudomonas putida in Complex with Uridine-diphosphate-2(n-acetylglucosaminyl) butyric acid, (2R)-2-(phosphonooxy)propanoic acid and Magnesium.
To Be Published
|
|
5XWB
 
 | | Crystal Structure of 5-Enolpyruvulshikimate-3-phosphate Synthase from a Psychrophilic Bacterium, Colwellia psychrerythraea | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase | | Authors: | Lee, J.H, Kim, H.J, Choi, J.M, Kim, D.-W, Seo, Y.-S. | | Deposit date: | 2017-06-29 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of 5-enolpyruvylshikimate-3-phosphate synthase from a psychrophilic bacterium, Colwellia psychrerythraea 34H.
Biochem. Biophys. Res. Commun., 492, 2017
|
|
2AA9
 
 | | EPSP synthase liganded with shikimate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 3-phosphoshikimate 1-carboxyvinyltransferase, FORMIC ACID | | Authors: | Priestman, M.A, Healy, M.L, Funke, T, Becker, A, Schonbrunn, E. | | Deposit date: | 2005-07-13 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular basis for the glyphosate-insensitivity of the reaction of 5-enolpyruvylshikimate 3-phosphate synthase with shikimate.
Febs Lett., 579, 2005
|
|
5BQ2
 
 | |
4R7U
 
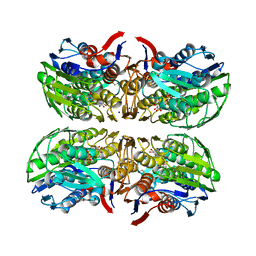 | | Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Vibrio cholerae in complex with substrate UDP-N-acetylglucosamine and the drug fosfomycin | | Descriptor: | SODIUM ION, TETRAETHYLENE GLYCOL, UDP-N-acetylglucosamine 1-carboxyvinyltransferase, ... | | Authors: | Nocek, B, Maltseva, N, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-28 | | Release date: | 2014-09-24 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Vibrio cholerae in complex with substrate UDP-N-acetylglucosamine and the drug fosfomycin
To be Published
|
|
8D84
 
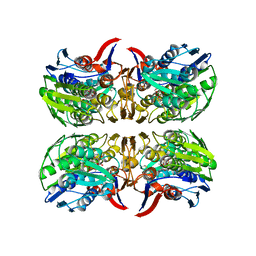 | | E. faecium MurAA in complex with UDP-N-acetylmuramic acid (UNAM) and a covalent adduct of PEP with Cys119 | | Descriptor: | (2R)-2-{[(2R,3R,4R,5S,6R)-3-(acetylamino)-2-{[(S)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-4-yl]oxy}propanoic acid, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Zhou, Y, Shamoo, Y. | | Deposit date: | 2022-06-07 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | E. faecium MurAA in complex with fosfomycin and UNAG
To Be Published
|
|
4ZND
 
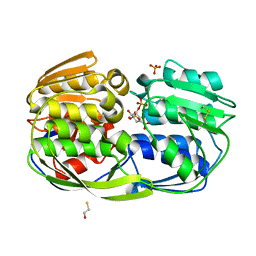 | | 2.55 Angstrom resolution structure of 3-phosphoshikimate 1-carboxyvinyltransferase (AroA) from Coxiella burnetii in complex with shikimate-3-phosphate, phosphate, and potassium | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, BETA-MERCAPTOETHANOL, PHOSPHATE ION, ... | | Authors: | Light, S.H, Minasov, G, Krishna, S.N, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-05-04 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | 2.55 Angstrom resolution structure of 3-phosphoshikimate 1-carboxyvinyltransferase (AroA) from Coxiella burnetii in complex with shikimate-3-phosphate, phosphate, and potassium
To Be Published
|
|
6WFM
 
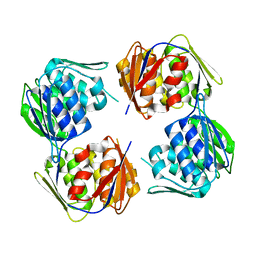 | |
1A2N
 
 | | STRUCTURE OF THE C115A MUTANT OF MURA COMPLEXED WITH THE FLUORINATED ANALOG OF THE REACTION TETRAHEDRAL INTERMEDIATE | | Descriptor: | UDP-N-ACETYLGLUCOSAMINE ENOLPYRUVYL TRANSFERASE, URIDINE-DIPHOSPHATE-2(N-ACETYLGLUCOSAMINYL-3-FLUORO-2-PHOSPHONOOXY)PROPIONIC ACID | | Authors: | Skarzynski, T. | | Deposit date: | 1998-01-06 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Stereochemical course of enzymatic enolpyruvyl transfer and catalytic conformation of the active site revealed by the crystal structure of the fluorinated analogue of the reaction tetrahedral intermediate bound to the active site of the C115A mutant of MurA
Biochemistry, 37, 1998
|
|
6NKJ
 
 | | 1.3 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Streptococcus pneumoniae in Complex with (2R)-2-(phosphonooxy)propanoic acid. | | Descriptor: | (2R)-2-(phosphonooxy)propanoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-01-07 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | 1.3 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Streptococcus pneumoniae in Complex with (2R)-2-(phosphonooxy)propanoic acid.
To Be Published
|
|
7PXY
 
 | |
7M0O
 
 | | DGT-28 EPSPS | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, PHOSPHATE ION, POTASSIUM ION | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2021-03-11 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Characterization of a Glyphosate-Tolerant Enzyme from Streptomyces svecius : A Distinct Class of 5-Enolpyruvylshikimate-3-phosphate Synthases.
J.Agric.Food Chem., 69, 2021
|
|
2YVW
 
 | |
3ZH3
 
 | |
2BJB
 
 | | Mycobacterium Tuberculosis Epsp Synthase In Unliganded State | | Descriptor: | 3-PHOSPHOSHIKIMATE 1-CARBOXYVINYLTRANSFERASE, ACETATE ION, SODIUM ION | | Authors: | Bourenkov, G.P, Kachalova, G.S, Strizhov, N, Bruning, M, Vagin, A, Bartunik, H.D. | | Deposit date: | 2005-02-01 | | Release date: | 2006-03-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mycobacterium Tuberculosis Epsp Synthase in Unliganded State
To be Published
|
|
3ZH4
 
 | |
2Z2C
 
 | | MURA inhibited by unag-cnicin adduct | | Descriptor: | 2-acetamido-3-O-[(2S,3S)-2-carboxy-3,4-dihydroxybutan-2-yl]-2-deoxy-alpha-D-glucopyranose, UDP-N-acetylglucosamine 1-carboxyvinyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Steinbach, A, Skarzynski, T, Scheidig, A.J, Klein, C.D. | | Deposit date: | 2007-05-17 | | Release date: | 2008-05-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Antibacterial Target Enzyme Mura Synthesizes its Own Non-Covalent Suicide Inhibitor from the Natural Product Cnicin
To be Published
|
|
2AAY
 
 | | EPSP synthase liganded with shikimate and glyphosate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 3-phosphoshikimate 1-carboxyvinyltransferase, FORMIC ACID, ... | | Authors: | Priestman, M.A, Healy, M.L, Funke, T, Becker, A. | | Deposit date: | 2005-07-14 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Molecular basis for the glyphosate-insensitivity of the reaction of 5-enolpyruvylshikimate 3-phosphate synthase with shikimate.
Febs Lett., 579, 2005
|
|
1X8T
 
 | | EPSPS liganded with the (R)-phosphonate analog of the tetrahedral reaction intermediate | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, FORMIC ACID, [3R-[3A,4A,5B(R*)]]-5-(1-CARBOXY-1-PHOSPHONOETHOXY)-4-HYDROXY-3-(PHOSPHONOOXY)-1-CYCLOHEXENE-1-CARBOXYLIC ACID | | Authors: | Priestman, M.A, Healy, M.L, Becker, A, Alberg, D.G, Bartlett, P.A, Schonbrunn, E. | | Deposit date: | 2004-08-18 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Interaction of phosphonate analogues of the tetrahedral reaction intermediate with 5-enolpyruvylshikimate-3-phosphate synthase in atomic detail.
Biochemistry, 44, 2005
|
|
1YBG
 
 | | MurA inhibited by a derivative of 5-sulfonoxy-anthranilic acid | | Descriptor: | N-METHYL-N-{2-[(2-NAPHTHYLSULFONYL)AMINO]-5-[(2-NAPHTHYLSULFONYL)OXY]BENZOYL}-L-ASPARTIC ACID, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Eschenburg, S, Priestman, M.A, Abdul-Latif, F.A, Delachaume, C, Fassy, F, Schonbrunn, E. | | Deposit date: | 2004-12-20 | | Release date: | 2005-02-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Novel Inhibitor That Suspends the Induced Fit Mechanism of UDP-N-acetylglucosamine Enolpyruvyl Transferase (MurA).
J.Biol.Chem., 280, 2005
|
|
1X8R
 
 | | EPSPS liganded with the (S)-phosphonate analog of the tetrahedral reaction intermediate | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, FORMIC ACID, [3R-[3A,4A,5B(S*)]]-5-(1-CARBOXY-1-PHOSPHONOETHOXY)-4-HYDROXY-3-(PHOSPHONOOXY)-1-CYCLOHEXENE-1-CARBOXYLIC ACID | | Authors: | Priestman, M.A, Healy, M.L, Becker, A, Alberg, D.G, Bartlett, P.A, Schonbrunn, E. | | Deposit date: | 2004-08-18 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Interaction of phosphonate analogues of the tetrahedral reaction intermediate with 5-enolpyruvylshikimate-3-phosphate synthase in atomic detail.
Biochemistry, 44, 2005
|
|
5U4H
 
 | | 1.05 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Acinetobacter baumannii in Covalently Bound Complex with (2R)-2-(phosphonooxy)propanoic Acid. | | Descriptor: | (2R)-2-(phosphonooxy)propanoic acid, FORMIC ACID, SODIUM ION, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-04 | | Release date: | 2016-12-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | 1.05 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Acinetobacter baumannii in Covalently Bound Complex with (2R)-2-(phosphonooxy)propanoic Acid.
To Be Published
|
|
5UJS
 
 | | 2.45 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Campylobacter jejuni. | | Descriptor: | CHLORIDE ION, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Stam, J, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-01-18 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | 2.45 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Campylobacter jejuni.
To Be Published
|
|
6Q03
 
 | | Crystal structure of MurA from Clostridium difficile in the presence of UDP-N-acetyl-alpha-D-muramic acid with modified Cys116 (S-[(1S)-1-carboxy-1-(phosphonooxy)ethyl]-L-cysteine) | | Descriptor: | (2R)-2-{[(2R,3R,4R,5S,6R)-3-(acetylamino)-2-{[(S)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-4-yl]oxy}propanoic acid, 1,2-ETHANEDIOL, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Dopkins, B.J, Call, C.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2019-08-01 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of MurA from Clostridium difficile in the presence of UDP-N-acetyl-alpha-D-muramic acid with modified Cys116 (S-[(1S)-1-carboxy-1-(phosphonooxy)ethyl]-L-cysteine)
To Be Published
|
|
6Q0A
 
 | | Crystal structure of MurA from Clostridium difficile, mutation C116D, n the presence of UDP-N-acetylmuramic acid | | Descriptor: | (2R)-2-{[(2R,3R,4R,5S,6R)-3-(acetylamino)-2-{[(S)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-4-yl]oxy}propanoic acid, 1,2-ETHANEDIOL, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Dopkins, B.J, Call, C.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2019-08-01 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of MurA from Clostridium difficile, mutation C116D, n the presence of UDP-N-acetylmuramic acid
To Be Published
|
|
