1SPI
 
 | | CRYSTAL STRUCTURE OF SPINACH CHLOROPLAST FRUCTOSE-1,6-BISPHOSPHATASE AT 2.8 ANGSTROMS RESOLUTION | | Descriptor: | FRUCTOSE 1,6-BISPHOSPHATASE | | Authors: | Villeret, V, Huang, S, Zhang, Y, Xue, Y, Lipscomb, W.N. | | Deposit date: | 1994-12-14 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of spinach chloroplast fructose-1,6-bisphosphatase at 2.8 A resolution.
Biochemistry, 34, 1995
|
|
1SPJ
 
 | | STRUCTURE OF MATURE HUMAN TISSUE KALLIKREIN (HUMAN KALLIKREIN 1 OR KLK1) AT 1.70 ANGSTROM RESOLUTION WITH VACANT ACTIVE SITE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, CALCIUM ION, ... | | Authors: | Laxmikanthan, G, Blaber, S.I, Bernett, M.J, Blaber, M. | | Deposit date: | 2004-03-16 | | Release date: | 2005-01-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.70 A X-ray structure of human apo kallikrein 1: Structural changes upon peptide inhibitor/substrate binding
Proteins, 58, 2005
|
|
1SPK
 
 | | Solution Structure of RSGI RUH-010, an SH3 Domain from Mouse cDNA | | Descriptor: | RIKEN cDNA 1300006M19 | | Authors: | Suzuki, Y, Abe, T, Hirota, H, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-17 | | Release date: | 2004-09-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of RSGI RUH-010, an SH3 Domain from Mouse cDNA
To be Published
|
|
1SPP
 
 | | THE CRYSTAL STRUCTURES OF TWO MEMBERS OF THE SPERMADHESIN FAMILY REVEAL THE FOLDING OF THE CUB DOMAIN | | Descriptor: | MAJOR SEMINAL PLASMA GLYCOPROTEIN PSP-I, MAJOR SEMINAL PLASMA GLYCOPROTEIN PSP-II | | Authors: | Romero, A, Romao, M.J, Varela, P.F, Kolln, I, Dias, J.M, Carvalho, A.L, Sanz, L, Topfer-Petersen, E, Calvete, J.J. | | Deposit date: | 1997-06-19 | | Release date: | 1998-06-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structures of two spermadhesins reveal the CUB domain fold.
Nat.Struct.Biol., 4, 1997
|
|
1SPQ
 
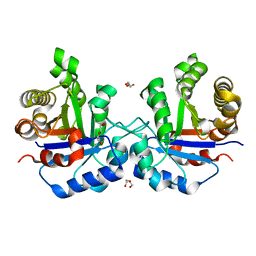 | | Understanding protein lids: Structural analysis of active hinge mutants in triosephosphate isomerase | | Descriptor: | DI(HYDROXYETHYL)ETHER, Triosephosphate isomerase | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-17 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
1SPR
 
 | |
1SPS
 
 | |
1SPU
 
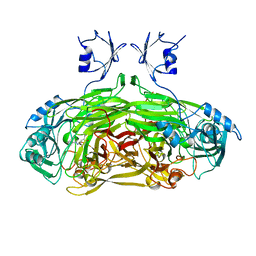 | | STRUCTURE OF OXIDOREDUCTASE | | Descriptor: | CALCIUM ION, COPPER (II) ION, COPPER AMINE OXIDASE | | Authors: | Wilmot, C.M, Phillips, S.E.V. | | Deposit date: | 1996-11-13 | | Release date: | 1997-03-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catalytic mechanism of the quinoenzyme amine oxidase from Escherichia coli: exploring the reductive half-reaction.
Biochemistry, 36, 1997
|
|
1SPV
 
 | | Crystal Structure of the Putative Phosphatase of Escherichia coli, Northeast Structural Genomoics Target ER58 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, putative polyprotein/phosphatase | | Authors: | Forouhar, F, Lee, I, Vorobiev, S.M, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-03-17 | | Release date: | 2004-04-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Putative Phosphatase of Escherichia coli, Northeast Structural Genomoics Target ER58
To be Published
|
|
1SPW
 
 | | Solution Structure of a Loop Truncated Mutant from D. gigas Rubredoxin, NMR | | Descriptor: | Rubredoxin | | Authors: | Pais, T.M, Lamosa, P, dos Santos, W, LeGall, J, Turner, D.L, Santos, H. | | Deposit date: | 2004-03-17 | | Release date: | 2005-03-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural determinants of protein stabilization by solutes: the importance of the hairpin loop in rubredoxins
FEBS J., 272, 2005
|
|
1SPX
 
 | | Crystal Structure of Glucose Dehydrogenase of Caenorhabditis Elegans in the Apo-Form | | Descriptor: | short-chain reductase family member (5L265) | | Authors: | Schormann, N, Zhou, J, McCombs, D, Bray, T, Symersky, J, Huang, W.-Y, Luan, C.-H, Gray, R, Luo, D, Arabashi, A, Bunzel, B, Nagy, L, Lu, S, Li, S, Lin, G, Zhang, Y, Qiu, S, Tsao, J, Luo, M, Carson, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-03-17 | | Release date: | 2004-03-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Glucose Dehydrogenase of Caenorhabditis Elegans in the Apo-Form: A Member of the SDR-Family
To be Published
|
|
1SPY
 
 | | REGULATORY DOMAIN OF HUMAN CARDIAC TROPONIN C IN THE CALCIUM-FREE STATE, NMR, 40 STRUCTURES | | Descriptor: | TROPONIN C | | Authors: | Spyracopoulos, L, Li, M.X, Sia, S.K, Gagne, S.M, Chandra, M, Solaro, R.J, Sykes, B.D. | | Deposit date: | 1997-07-14 | | Release date: | 1998-09-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Calcium-induced structural transition in the regulatory domain of human cardiac troponin C.
Biochemistry, 36, 1997
|
|
1SQ0
 
 | | Crystal Structure of the Complex of the Wild-type Von Willebrand Factor A1 domain and Glycoprotein Ib alpha at 2.6 Angstrom Resolution | | Descriptor: | Platelet glycoprotein Ib alpha chain (Glycoprotein Ibalpha) (GP-Ib alpha) (GPIbA) (GPIb-alpha) (CD42B-alpha) (CD42B) [Contains: Glycocalicin], Von Willebrand factor (vWF) [Contains: Von Willebrand antigen II] | | Authors: | Dumas, J.J, Kumar, R, McDonagh, T, Sullivan, F, Stahl, M.L, Somers, W.S, Mosyak, L. | | Deposit date: | 2004-03-17 | | Release date: | 2004-04-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the wild-type von Willebrand factor A1-glycoprotein Ibalpha complex reveals conformation differences with a complex bearing von Willebrand disease mutations
J.Biol.Chem., 279, 2004
|
|
1SQ1
 
 | | Crystal Structure of the Chorismate Synthase from Campylobacter jejuni, Northeast Structural Genomics Target BR19 | | Descriptor: | Chorismate synthase, SULFATE ION | | Authors: | Forouhar, F, Lee, I, Vorobiev, S.M, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-03-17 | | Release date: | 2004-04-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Chorismate Synthase from Campylobacter jejuni, Northeast Structural Genomics Target BR19
To be Published
|
|
1SQ2
 
 | | Crystal Structure Analysis of the Nurse Shark New Antigen Receptor (NAR) Variable Domain in Complex With Lysozyme | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Stanfield, R.L, Dooley, H, Flajnik, M.F, Wilson, I.A. | | Deposit date: | 2004-03-17 | | Release date: | 2004-08-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of a shark single-domain antibody V region in complex with lysozyme.
Science, 305, 2004
|
|
1SQ3
 
 | | Crystal structures of a novel open pore ferritin from the hyperthermophilic Archaeon Archaeoglobus fulgidus. | | Descriptor: | FE (III) ION, ferritin | | Authors: | Johnson, E, Cascio, D, Michael, S, Schroder, I. | | Deposit date: | 2004-03-17 | | Release date: | 2005-04-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of a tetrahedral open pore ferritin from the hyperthermophilic archaeon Archaeoglobus fulgidus.
Structure, 13, 2005
|
|
1SQ4
 
 | | Crystal Structure of the Putative Glyoxylate Induced Protein from Pseudomonas aeruginosa, Northeast Structural Genomics Target PaR14 | | Descriptor: | Glyoxylate-induced Protein, THIOCYANATE ION | | Authors: | Forouhar, F, Chen, Y, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-03-17 | | Release date: | 2004-04-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Putative Glyoxylate Induced Protein from Pseudomonas aeruginosa, Northeast Structural Genomics Target PaR14
To be Published
|
|
1SQ5
 
 | | Crystal Structure of E. coli Pantothenate kinase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PANTOTHENOIC ACID, Pantothenate kinase | | Authors: | Ivey, R.A, Zhang, Y.-M, Virga, K.G, Hevener, K, Lee, R.E, Rock, C.O, Jackowski, S, Park, H.-W. | | Deposit date: | 2004-03-17 | | Release date: | 2004-09-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of the pantothenate kinase.ADP.pantothenate ternary complex reveals the relationship between the binding sites for substrate, allosteric regulator, and antimetabolites.
J.Biol.Chem., 279, 2004
|
|
1SQ6
 
 | |
1SQ7
 
 | | Understanding protein lids: Structural analysis of active hinge mutants in triosephosphate isomerase | | Descriptor: | Triosephosphate isomerase | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-18 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
1SQ8
 
 | |
1SQ9
 
 | |
1SQA
 
 | | Substituted 2-Naphthamidine Inhibitors of Urokinase | | Descriptor: | 6-[(Z)-AMINO(IMINO)METHYL]-N-[4-(AMINOMETHYL)PHENYL]-4-(PYRIMIDIN-2-YLAMINO)-2-NAPHTHAMIDE, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Wendt, M.D, Geyer, A, McClellan, W.J, Rockway, T.W, Weitzberg, M, Zhao, X, Stewart, K, Nienaber, V, Klinghofer, V, Giranda, V.L. | | Deposit date: | 2004-03-18 | | Release date: | 2004-04-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Interaction with the S1beta-pocket of urokinase: 8-heterocycle substituted and 6,8-disubstituted 2-naphthamidine urokinase inhibitors.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
1SQB
 
 | | Crystal Structure Analysis of Bovine Bc1 with Azoxystrobin | | Descriptor: | Cytochrome b, Cytochrome c1, heme protein, ... | | Authors: | Esser, L, Quinn, B, Li, Y.F, Zhang, M, Elberry, M, Yu, L, Yu, C.A, Xia, D. | | Deposit date: | 2004-03-18 | | Release date: | 2004-09-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystallographic studies of quinol oxidation site inhibitors: a modified classification of inhibitors for the cytochrome bc(1) complex
J.Mol.Biol., 341, 2004
|
|
1SQC
 
 | |
