1NYX
 
 | | Ligand binding domain of the human peroxisome proliferator activated receptor gamma in complex with an agonist | | Descriptor: | (2S)-2-ETHOXY-3-{4-[2-(10H-PHENOXAZIN-10-YL)ETHOXY]PHENYL}PROPANOIC ACID, peroxisome proliferator activated receptor gamma | | Authors: | Ebdrup, S, Pettersson, I, Rasmussen, H.B, Deussen, H.-J, Frost Jensen, A, Mortensen, S.B, Fleckner, J, Pridal, L, Nygaard, L, Sauerberg, P. | | Deposit date: | 2003-02-14 | | Release date: | 2003-07-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Synthesis and biological and structural characterization of the dual-acting peroxisome proliferator-activated receptor alpha/gamma agonist ragaglitazar
J.MED.CHEM., 46, 2003
|
|
1NYY
 
 | | Crystal Structure of the complex between M182T mutant of TEM-1 and a boronic acid inhibitor (105) | | Descriptor: | Beta-lactamase TEM, N-[5-METHYL-3-O-TOLYL-ISOXAZOLE-4-CARBOXYLIC ACID AMIDE] BORONIC ACID | | Authors: | Wang, X, Minasov, G, Blazquez, J, Caselli, E, Prati, F, Shoichet, B.K. | | Deposit date: | 2003-02-14 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Recognition and resistance in TEM beta-lactamase
Biochemistry, 42, 2003
|
|
1NZ0
 
 | | RNASE P PROTEIN FROM THERMOTOGA MARITIMA | | Descriptor: | Ribonuclease P protein component, SULFATE ION | | Authors: | Kazantsev, A.V, Krivenko, A.A, Harrington, D.J, Carter, R.J, Holbrook, S.R, Adams, P.D, Pace, N.R, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2003-02-14 | | Release date: | 2003-06-24 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High-resolution structure of RNase P protein from Thermotoga maritima.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NZ1
 
 | | Solution structure of the S. cerevisiae U6 Intramolecular stem-loop containing an SP phosphorothioate at nucleotide U80 | | Descriptor: | SP U6 Intramolecular Stem-Loop RNA | | Authors: | Reiter, N.J, Nikstad, L.J, Allman, A.M, Johnson, R.J, Butcher, S.E. | | Deposit date: | 2003-02-14 | | Release date: | 2003-05-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the U6 RNA intramolecular stem-loop harboring an
S(P)-phosphorothioate modification.
RNA, 9, 2003
|
|
1NZ2
 
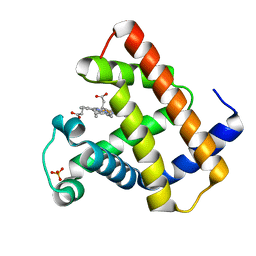 | | K45E Variant of Horse Heart Myoglobin | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Hunter, C.L, Maurus, R, Mauk, M.R, Lee, H, Raven, E.L, Tong, H, Nguyen, N, Smith, M, Brayer, G.D, Mauk, A.G. | | Deposit date: | 2003-02-15 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Introduction and characterization of a functionally linked metal ion binding site at
the exposed heme edge of myoglobin
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NZ3
 
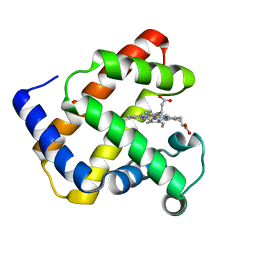 | | K45E-K63E Variant of Horse Heart Myoglobin | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hunter, C.L, Maurus, R, Mauk, M.R, Lee, H, Raven, E.L, Tong, H, Nguyen, N, Smith, S, Brayer, G.D, Mauk, A.G. | | Deposit date: | 2003-02-15 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Introduction and characterization of a functionally linked metal ion binding site at the
exposed heme edge of myoglobin
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NZ4
 
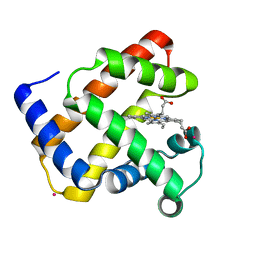 | | The horse heart myoglobin variant K45E/K63E complexed with Cadmium | | Descriptor: | CADMIUM ION, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hunter, C.L, Maurus, R, Mauk, M.R, Lee, H, Raven, E.L, Tong, H, Nguyen, N, Smith, S, Brayer, G.D, Mauk, A.G. | | Deposit date: | 2003-02-15 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Introduction and characterization of a functionally linked metal ion binding site at
the exposed heme edge of myoglobin
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NZ5
 
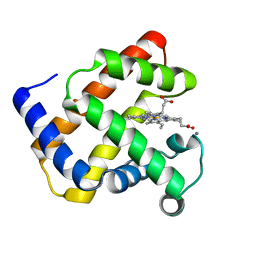 | | The Horse heart myoglobin variant K45E/K63E complexed with Manganese | | Descriptor: | MANGANESE (II) ION, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hunter, C.L, Maurus, R, Mauk, M.R, Lee, H, Raven, E.L, Tong, H, Nguyen, N, Smith, S, Brayer, G.D, Mauk, A.G. | | Deposit date: | 2003-02-15 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Introduction and characterization of a functionally linked metal ion binding site at
the exposed heme edge of myoglobin
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NZ6
 
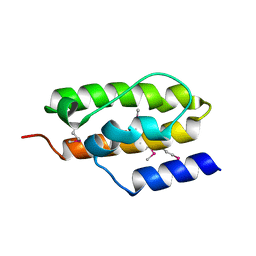 | | Crystal Structure of Auxilin J-Domain | | Descriptor: | Auxilin | | Authors: | Jiang, J, Taylor, A.B, Prasad, K, Ishikawa-Brush, Y, Hart, P.J, Lafer, E.M, Sousa, R. | | Deposit date: | 2003-02-16 | | Release date: | 2003-04-22 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-function analysis of the auxilin J-domain reveals an extended Hsc70 interaction interface.
Biochemistry, 42, 2003
|
|
1NZ7
 
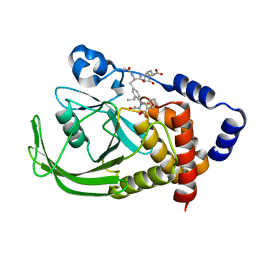 | | POTENT, SELECTIVE INHIBITORS OF PROTEIN TYROSINE PHOSPHATASE 1B USING A SECOND PHOSPHOTYROSINE BINDING SITE, complexed with compound 19. | | Descriptor: | 2-[(4-{2-ACETYLAMINO-2-[4-(1-CARBOXY-3-METHYLSULFANYL-PROPYLCARBAMOYL)-BUTYLCARBAMOYL]-ETHYL}-2-ETHYL-PHENYL)-OXALYL-AM INO]-BENZOIC ACID, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Xin, Z, Oost, T.K, Abad-Zapatero, C, Hajduk, P.J, Pei, Z, Szczepankiewicz, B.G, Hutchins, C.W, Ballaron, S.J, Stashko, M.A, Lubben, T, Trevillyan, J.M, Jirousek, M.R, Liu, G. | | Deposit date: | 2003-02-16 | | Release date: | 2003-05-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Potent, Selective Inhibitors of Protein Tyrosine Phosphatase 1B
BIOORG.MED.CHEM.LETT., 13, 2003
|
|
1NZ8
 
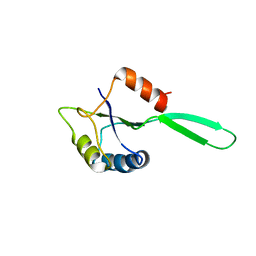 | | Solution Structure of the N-utilization substance G (NusG) N-terminal (NGN) domain from Thermus thermophilus | | Descriptor: | TRANSCRIPTION ANTITERMINATION PROTEIN NUSG | | Authors: | Reay, P, Yamasaki, K, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-17 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and sequence comparisons arising from the solution structure of the transcription elongation factor NusG from Thermus thermophilus
Proteins, 56, 2004
|
|
1NZ9
 
 | | Solution Structure of the N-utilization substance G (NusG) C-terminal (NGC) domain from Thermus thermophilus | | Descriptor: | TRANSCRIPTION ANTITERMINATION PROTEIN NUSG | | Authors: | Reay, P, Yamasaki, K, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-17 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and sequence comparisons arising from the solution structure of the transcription elongation factor NusG from Thermus thermophilus
Proteins, 56, 2004
|
|
1NZA
 
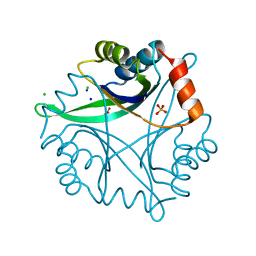 | | Divalent cation tolerance protein (Cut A1) from thermus thermophilus HB8 | | Descriptor: | CHLORIDE ION, Divalent cation tolerance protein, GLYCEROL, ... | | Authors: | Bagautdinov, B, Miyano, M, Tahirov, T.H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-17 | | Release date: | 2003-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structures of the CutA1 proteins from Thermus thermophilus and Pyrococcus horikoshii: characterization of metal-binding sites and metal-induced assembly.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
1NZB
 
 | | Crystal structure of wild type Cre recombinase-loxP synapse | | Descriptor: | Cre recombinase, IODIDE ION, MAGNESIUM ION, ... | | Authors: | Ennifar, E, Meyer, J.E.W, Buchholz, F, Stewart, A.F, Suck, D. | | Deposit date: | 2003-02-17 | | Release date: | 2003-09-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of a wild-type Cre recombinase-loxP synapse reveals a novel spacer conformation suggesting an alternative mechanism for DNA cleavage activation
Nucleic Acids Res., 31, 2003
|
|
1NZC
 
 | | The high resolution structures of RmlC from Streptococcus suis in complex with dTDP-D-xylose | | Descriptor: | NICKEL (II) ION, THYMIDINE-5'-DIPHOSPHO-BETA-D-XYLOSE, dTDP-6-deoxy-D-xylo-4-hexulose 3,5-epimerase | | Authors: | Dong, C, Major, L.L, Allen, A, Blankenfeldt, W, Maskell, D, Naismith, J.H. | | Deposit date: | 2003-02-17 | | Release date: | 2003-06-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-Resolution Structures of RmlC from Streptococcus suis in Complex with Substrate Analogs Locate the Active Site of This Class of Enzyme
Structure, 11, 2003
|
|
1NZD
 
 | | T4 phage BGT-D100A mutant in complex with UDP-glucose: Form I | | Descriptor: | CHLORIDE ION, DNA beta-glycosyltransferase, GLYCEROL, ... | | Authors: | Lariviere, L, Morera, S. | | Deposit date: | 2003-02-17 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the T4 phage beta-glucosyltransferase and the D100A mutant in complex with UDP-glucose: glucose binding and identification of the catalytic base for a direct displacement mechanism
J.Mol.Biol., 330, 2003
|
|
1NZE
 
 | | Crystal structure of PsbQ polypeptide of photosystem II from higher plants | | Descriptor: | Oxygen-evolving enhancer protein 3, ZINC ION | | Authors: | Calderone, V, Trabucco, M, Vujicic, A, Battistutta, R, Giacometti, G.M, Andreucci, F, Barbato, R, Zanotti, G. | | Deposit date: | 2003-02-17 | | Release date: | 2003-08-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the PsbQ protein of photosystem II from higher plants
Embo Rep., 4, 2003
|
|
1NZF
 
 | | T4 phage BGT-D100A mutant in complex with UDP-glucose: Form II | | Descriptor: | CHLORIDE ION, DNA beta-glycosyltransferase, GLYCEROL, ... | | Authors: | Lariviere, L, Morera, S. | | Deposit date: | 2003-02-17 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the T4 phage beta-glucosyltransferase and the D100A mutant in complex with
UDP-glucose: glucose binding and identification of the catalytic base for a direct displacement mechanism.
J.Mol.Biol., 330, 2003
|
|
1NZG
 
 | | Crystal structure of A-DNA decamer GCGTA(3ME)ACGC, with a modified 5-methyluridine | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*(3ME)P*AP*CP*GP*C)-3' | | Authors: | Prhavc, M, Prakash, T.P, Minasov, G, Egli, M, Manoharan, M. | | Deposit date: | 2003-02-17 | | Release date: | 2003-08-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 2'-O-[2-[2-(N,N-dimethylamino)ethoxy]ethyl] modified oligonucleotides: symbiosis of charge interaction factors and stereoelectronic effects
Org.Lett., 5, 2003
|
|
1NZI
 
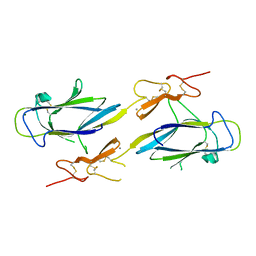 | | Crystal Structure of the CUB1-EGF Interaction Domain of Complement Protease C1s | | Descriptor: | CALCIUM ION, Complement C1s component, MAGNESIUM ION | | Authors: | Gregory, L.A, Thielens, N.M, Arlaud, G.J, Fontecilla-Camps, J.C, Gaboriaud, C. | | Deposit date: | 2003-02-18 | | Release date: | 2003-06-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray structure of the Ca2+-binding interaction domain of C1s. Insights into the assembly of the C1 complex of complement
J.Biol.Chem., 278, 2003
|
|
1NZJ
 
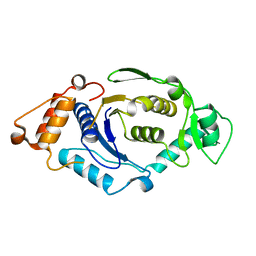 | | Crystal Structure and Activity Studies of Escherichia Coli Yadb ORF | | Descriptor: | Hypothetical protein yadB, ZINC ION | | Authors: | Campanacci, V, Kern, D.Y, Becker, H.D, Spinelli, S, Valencia, C, Vincentelli, R, Pagot, F, Bignon, C, Giege, R, Cambillau, C. | | Deposit date: | 2003-02-18 | | Release date: | 2004-04-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Escherichia coli YadB gene product reveals a novel aminoacyl-tRNA synthetase like activity.
J.Mol.Biol., 337, 2004
|
|
1NZK
 
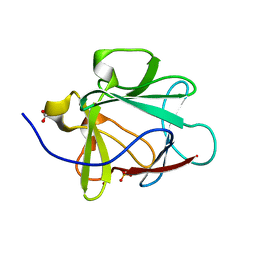 | | Crystal Structure of a Multiple Mutant (L44F, L73V, V109L, L111I, C117V) of Human Acidic Fibroblast Growth Factor | | Descriptor: | Acidic Fibroblast Growth Factor, FORMIC ACID, SULFATE ION | | Authors: | Brych, S.R, Kim, J, Logan, T.M, Blaber, M. | | Deposit date: | 2003-02-18 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Accommodation of a highly symmetric core within a symmetric protein superfold.
Protein Sci., 12, 2003
|
|
1NZL
 
 | |
1NZM
 
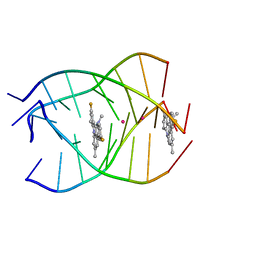 | | NMR structure of the parallel-stranded DNA quadruplex d(TTAGGGT)4 complexed with the telomerase inhibitor RHPS4 | | Descriptor: | 3,11-DIFLUORO-6,8,13-TRIMETHYL-8H-QUINO[4,3,2-KL]ACRIDIN-13-IUM, 5'-D(*TP*TP*AP*GP*GP*GP*T)-3', POTASSIUM ION | | Authors: | Gavathiotis, E, Heald, R.A, Stevens, M.F.G, Searle, M.S. | | Deposit date: | 2003-02-18 | | Release date: | 2003-11-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Drug Recognition and Stabilisation of the Parallel-stranded DNA Quadruplex d(TTAGGGT)4 Containing the Human Telomeric Repeat
J.Mol.Biol., 334, 2003
|
|
1NZN
 
 | |
