6XCR
 
 | | NMR structure of Ost4 in DPC micelles | | Descriptor: | Oligosaccharyltransferase | | Authors: | Chaudhary, B.P. | | Deposit date: | 2020-06-09 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR and MD simulations reveal the impact of the V23D mutation on the function of yeast oligosaccharyltransferase subunit Ost4.
Glycobiology, 31, 2021
|
|
6XCU
 
 | |
5MEI
 
 | | Crystal structure of Agelastatin A bound to the 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | McClary, B, Zinshteyn, B, Meyer, M, Jouanneau, M, Pellegrino, S, Yusupova, G, Schuller, A, Reyes, J.C.P, Lu, J, Luo, C, Dang, Y, Romo, D, Yusupov, M, Green, R, Liu, J.O. | | Deposit date: | 2016-11-15 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Inhibition of Eukaryotic Translation by the Antitumor Natural Product Agelastatin A.
Cell Chem Biol, 24, 2017
|
|
1QAX
 
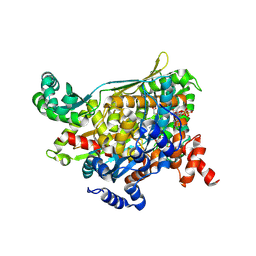 | | TERNARY COMPLEX OF PSEUDOMONAS MEVALONII HMG-COA REDUCTASE WITH HMG-COA AND NAD+ | | Descriptor: | 3-HYDROXY-3-METHYLGLUTARYL-COENZYME A, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROTEIN (3-HYDROXY-3-METHYLGLUTARYL-COENZYME A REDUCTASE) | | Authors: | Tabernero, L, Bochar, D.A, Rodwell, V.W, Stauffacher, C.V. | | Deposit date: | 1999-04-06 | | Release date: | 1999-06-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Substrate-induced closure of the flap domain in the ternary complex structures provides insights into the mechanism of catalysis by 3-hydroxy-3-methylglutaryl-CoA reductase.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
5U1F
 
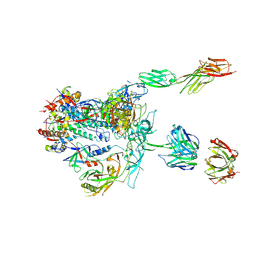 | | Initial contact of HIV-1 Env with CD4: Cryo-EM structure of BG505 DS-SOSIP trimer in complex with CD4 and antibody PGT145 | | Descriptor: | BG505 DS-SOSIP gp120, BG505 SOSIP gp41, PGT145 heavy chain, ... | | Authors: | Acharya, P, Kwong, P.D, Potter, C.S, Carragher, B. | | Deposit date: | 2016-11-28 | | Release date: | 2017-02-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Quaternary contact in the initial interaction of CD4 with the HIV-1 envelope trimer.
Nat. Struct. Mol. Biol., 24, 2017
|
|
1UNS
 
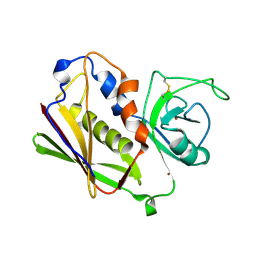 | | IDENTIFICATION OF A SECONDARY ZINC-BINDING SITE IN STAPHYLOCOCCAL ENTEROTOXIN C2: IMPLICATIONS FOR SUPERANTIGEN RECOGNITION | | Descriptor: | ENTEROTOXIN TYPE C-2, ZINC ION | | Authors: | Papageorgiou, A.C, Baker, M.D, McLeod, J.D, Goda, S, Sansom, D.M, Tranter, H.S, Acharya, K.R. | | Deposit date: | 2003-09-15 | | Release date: | 2003-11-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of a Secondary Zinc-Binding Site in Staphylococcal Enterotoxin C2: Implications for Superantigen Recognition
J.Biol.Chem., 279, 2004
|
|
1PLQ
 
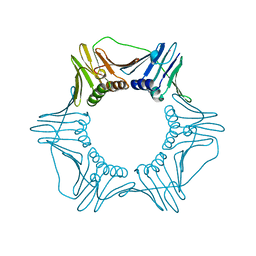 | | CRYSTAL STRUCTURE OF THE EUKARYOTIC DNA POLYMERASE PROCESSIVITY FACTOR PCNA | | Descriptor: | MERCURY (II) ION, PROLIFERATING CELL NUCLEAR ANTIGEN (PCNA) | | Authors: | Krishna, T.S.R, Kong, X.-P, Gary, S, Burgers, P.M, Kuriyan, J. | | Deposit date: | 1995-01-02 | | Release date: | 1995-03-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA.
Cell(Cambridge,Mass.), 79, 1994
|
|
1PLR
 
 | | CRYSTAL STRUCTURE OF THE EUKARYOTIC DNA POLYMERASE PROCESSIVITY FACTOR PCNA | | Descriptor: | PROLIFERATING CELL NUCLEAR ANTIGEN (PCNA) | | Authors: | Krishna, T.S.R, Kong, X.-P, Gary, S, Burgers, P.M, Kuriyan, J. | | Deposit date: | 1995-01-02 | | Release date: | 1995-03-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA.
Cell(Cambridge,Mass.), 79, 1994
|
|
5Z4W
 
 | | Crystal structure of signalling protein from buffalo (SPB-40) with an altered conformation of Trp78 at 1.79 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1, ... | | Authors: | Singh, P.K, Chaudhary, A, Tyagi, T.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2018-01-15 | | Release date: | 2018-02-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A glycoprotein from mammary gland secreted during involution promotes apoptosis: Structural and biological studies.
Arch. Biochem. Biophys., 644, 2018
|
|
4I6A
 
 | | 3-hydroxy-3-methylglutaryl (HMG) Coenzyme A reductase from Pseudomonas mevalonii complexed with HMG-CoA | | Descriptor: | 3-HYDROXY-3-METHYLGLUTARYL-COENZYME A, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, SULFATE ION | | Authors: | Steussy, C.N, Stauffacher, C.V, Schmidt, T, Burgner II, J.W, Rodwell, V.W, Wrensford, L.V, Critchelow, C.J, Min, J. | | Deposit date: | 2012-11-29 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Novel Role for Coenzyme A during Hydride Transfer in 3-Hydroxy-3-methylglutaryl-coenzyme A Reductase.
Biochemistry, 52, 2013
|
|
5Z3S
 
 | | Crystal structure of butanol modified signaling protein from buffalo (SPB-40) at 1.65 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-BUTANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, P.K, Chaudhary, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2018-01-08 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A glycoprotein from mammary gland secreted during involution promotes apoptosis: Structural and biological studies.
Arch. Biochem. Biophys., 644, 2018
|
|
5Z05
 
 | | Crystal structure of signalling protein from buffalo (SPB-40) with an acetone induced conformation of Trp78 at 1.49 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETONE, ... | | Authors: | Singh, P.K, Chaudhary, A, Tyagi, T.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2017-12-18 | | Release date: | 2018-01-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A glycoprotein from mammary gland secreted during involution promotes apoptosis: Structural and biological studies
Arch. Biochem. Biophys., 644, 2018
|
|
2VXM
 
 | | Screening a Limited Structure-based Library Identifies UDP-GalNAc- Specific Mutants of alpha-1,3 Galactosyltransferase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE | | Authors: | Tumbale, P, Jamaluddin, H, Thiyagarajan, N, Acharya, K.R, Brew, K. | | Deposit date: | 2008-07-07 | | Release date: | 2008-09-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Screening a Limited Structure-Based Library Identifies Udp-Galnac-Specific Mutants of {Alpha}-1,3-Galactosyltransferase.
Glycobiology, 18, 2008
|
|
2VXL
 
 | | Screening a Limited Structure-based Library Identifies UDP-GalNAc- Specific Mutants of alpha-1,3 Galactosyltransferase | | Descriptor: | MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE | | Authors: | Tumbale, P, Jamaluddin, H, Thiyagarajan, N, Acharya, K.R, Brew, K. | | Deposit date: | 2008-07-07 | | Release date: | 2008-09-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Screening a Limited Structure-Based Library Identifies Udp-Galnac-Specific Mutants of {Alpha}-1,3-Galactosyltransferase.
Glycobiology, 18, 2008
|
|
2YB9
 
 | | Crystal Structure of Human Neutral Endopeptidase complexed with a heteroarylalanine diacid. | | Descriptor: | HETEROARYLALANINE 5-PHENYL OXAZOLE, NEPRILYSIN, ZINC ION | | Authors: | Glossop, M.S, Bazin, R.J, Dack, K.N, Done, S, Fox, D.N.A, MacDonald, G.A, Mills, M, Owen, D.R, Phillips, C, Reeves, K.A, Ringer, T.J, Strang, R.S, Watson, C.A.L. | | Deposit date: | 2011-03-02 | | Release date: | 2011-05-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis and Evaluation of Heteroarylalanine Diacids as Potent and Selective Neutral Endopeptidase Inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3GWJ
 
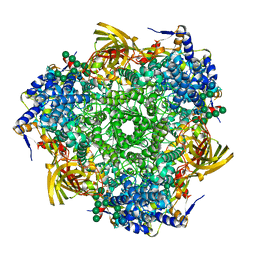 | | Crystal structure of Antheraea pernyi arylphorin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Arylphorin, FORMIC ACID, ... | | Authors: | Ryu, K.S, Lee, J.O, Kwon, T.H, Kim, S. | | Deposit date: | 2009-04-01 | | Release date: | 2009-05-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The presence of monoglucosylated N196-glycan is important for the structural stability of storage protein, arylphorin
Biochem.J., 421, 2009
|
|
1EIF
 
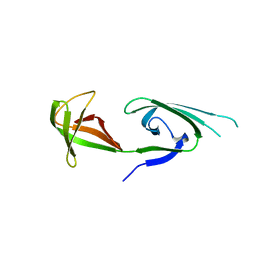 | | EUKARYOTIC TRANSLATION INITIATION FACTOR 5A FROM METHANOCOCCUS JANNASCHII | | Descriptor: | EUKARYOTIC TRANSLATION INITIATION FACTOR 5A | | Authors: | Kim, K.K, Hung, L.W, Yokota, H, Kim, R, Kim, S.H. | | Deposit date: | 1998-07-29 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of eukaryotic translation initiation factor 5A from Methanococcus jannaschii at 1.8 A resolution.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
4Z0M
 
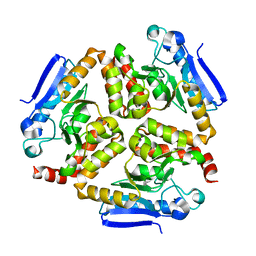 | | EchA5 Mycobacterium tuberculosis | | Descriptor: | Enoyl-CoA hydratase | | Authors: | Chaudhary, S, Gokhale, R.S. | | Deposit date: | 2015-03-26 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Unsaturated Lipid Assimilation by Mycobacteria Requires Auxiliary cis-trans Enoyl CoA Isomerase
Chem.Biol., 22, 2015
|
|
1CNT
 
 | | CILIARY NEUROTROPHIC FACTOR | | Descriptor: | CILIARY NEUROTROPHIC FACTOR, SULFATE ION, YTTERBIUM (III) ION | | Authors: | Mcdonald, N.Q, Panayotatos, N, Hendrickson, W.A. | | Deposit date: | 1996-06-06 | | Release date: | 1997-03-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of dimeric human ciliary neurotrophic factor determined by MAD phasing.
EMBO J., 14, 1995
|
|
4CYR
 
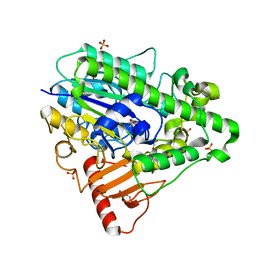 | | G4 mutant of PAS, arylsulfatase from Pseudomonas Aeruginosa | | Descriptor: | ARYLSULFATASE, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Miton, C.M, Jonas, S, Mohammed, M.F, Fischer, G, Loo, B.v, Kintses, B, Hyvonen, M, Tokuriki, N, Hollfelder, F. | | Deposit date: | 2014-04-14 | | Release date: | 2015-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Evolutionary repurposing of a sulfatase: A new Michaelis complex leads to efficient transition state charge offset.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4CXK
 
 | | G9 mutant of PAS, arylsulfatase from Pseudomonas Aeruginosa | | Descriptor: | ARYLSULFATASE, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Miton, C.M, Jonas, S, Mohammed, M.F, Fischer, G, Loo, B.v, Kintses, B, Hyvonen, M, Tokuriki, N, Hollfelder, F. | | Deposit date: | 2014-04-07 | | Release date: | 2015-04-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Evolutionary repurposing of a sulfatase: A new Michaelis complex leads to efficient transition state charge offset.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4CXS
 
 | | G4 mutant of PAS, arylsulfatase from Pseudomonas aeruginosa, in complex with Phenylphosphonic acid | | Descriptor: | ARYLSULFATASE, CALCIUM ION, SULFATE ION, ... | | Authors: | Miton, C.M, Jonas, S, Mohammed, M.F, Fischer, G, Loo, B.v, Kintses, B, Hyvonen, M, Tokuriki, N, Hollfelder, F. | | Deposit date: | 2014-04-08 | | Release date: | 2015-05-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolutionary repurposing of a sulfatase: A new Michaelis complex leads to efficient transition state charge offset.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3HDH
 
 | | PIG HEART SHORT CHAIN L-3-HYDROXYACYL COA DEHYDROGENASE REVISITED: SEQUENCE ANALYSIS AND CRYSTAL STRUCTURE DETERMINATION | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROTEIN (L-3-HYDROXYACYL COA DEHYDROGENASE) | | Authors: | Barycki, J.J, O'Brien, L.K, Birktoft, J.J, Strauss, A.W, Banaszak, L.J. | | Deposit date: | 1999-04-13 | | Release date: | 1999-10-08 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Pig heart short chain L-3-hydroxyacyl-CoA dehydrogenase revisited: sequence analysis and crystal structure determination.
Protein Sci., 8, 1999
|
|
3MPI
 
 | | Structure of the glutaryl-coenzyme A dehydrogenase glutaryl-CoA complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Glutaryl-CoA dehydrogenase, glutaryl-coenzyme A | | Authors: | Wischgoll, S, Warkentin, E, Boll, M, Ermler, U. | | Deposit date: | 2010-04-27 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for promoting and preventing decarboxylation in glutaryl-coenzyme a dehydrogenases.
Biochemistry, 49, 2010
|
|
2R0N
 
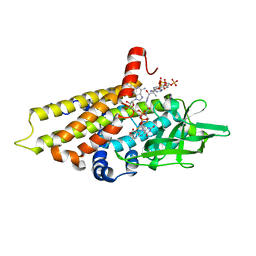 | | The effect of a Glu370Asp mutation in Glutaryl-CoA Dehydrogenase on Proton Transfer to the Dienolate Intermediate | | Descriptor: | 3-thiaglutaryl-CoA, FLAVIN-ADENINE DINUCLEOTIDE, Glutaryl-CoA dehydrogenase | | Authors: | Rao, K.S, Albro, M, Fu, Z, Narayanan, B, Baddam, S, Lee, H.J, Kim, J.J, Frerman, F.E. | | Deposit date: | 2007-08-20 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The effect of a Glu370Asp mutation in glutaryl-CoA dehydrogenase on proton transfer to the dienolate intermediate.
Biochemistry, 46, 2007
|
|
