9LDB
 
 | | DESIGN AND SYNTHESIS OF NEW ENZYMES BASED ON THE LACTATE DEHYDROGENASE FRAMEWORK | | Descriptor: | LACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, OXAMIC ACID, ... | | Authors: | Dunn, C.R, Holbrook, J.J, Muirhead, H. | | Deposit date: | 1991-11-26 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design and synthesis of new enzymes based on the lactate dehydrogenase framework.
Philos.Trans.R.Soc.London,Ser.B, 332, 1991
|
|
1TI3
 
 | | Solution structure of the Thioredoxin h1 from poplar, a CPPC active site variant | | Descriptor: | thioredoxin H | | Authors: | Coudevylle, N, Thureau, A, Hemmerlin, C, Gelhaye, E, Jacquot, J.P, Cung, M.T. | | Deposit date: | 2004-06-02 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a natural CPPC active site variant, the reduced form of thioredoxin h1 from poplar.
Biochemistry, 44, 2005
|
|
8PII
 
 | | VHH Z70 mutant 3 in interaction with PHF6 Tau peptide | | Descriptor: | Microtubule-associated protein tau, VHH Z70 Mutant 3 | | Authors: | Dupre, E, Mortelecque, J, NGuyen, M, Hanoulle, X, Landrieu, I. | | Deposit date: | 2023-06-21 | | Release date: | 2023-07-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A selection and optimization strategy for single-domain antibodies targeting the PHF6 linear peptide within the tau intrinsically disordered protein.
J.Biol.Chem., 300, 2024
|
|
8P60
 
 | | Spraguea lophii ribosome dimer | | Descriptor: | 40S Ribosomal protein S19, 40S ribosomal protein S0, 40S ribosomal protein S1, ... | | Authors: | Gil Diez, P, McLaren, M, Isupov, M.N, Daum, B, Conners, R, Williams, B. | | Deposit date: | 2023-05-24 | | Release date: | 2023-06-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (14.3 Å) | | Cite: | CryoEM reveals that ribosomes in microsporidian spores are locked in a dimeric hibernating state.
Nat Microbiol, 8, 2023
|
|
5IXE
 
 | | 1.75A RESOLUTION STRUCTURE OF 5-Fluoroindole BOUND BETA-GLYCOSIDASE (W33G) FROM SULFOLOBUS SOLFATARICUS | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-fluoro-1H-indole, Beta-galactosidase, ... | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Budiardjo, S.J, Karanicolas, J. | | Deposit date: | 2016-03-23 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Full and Partial Agonism of a Designed Enzyme Switch.
ACS Synth Biol, 5, 2016
|
|
1BVX
 
 | | THE 1.8 A STRUCTURE OF GEL GROWN TETRAGONAL HEN EGG WHITE LYSOZYME | | Descriptor: | PROTEIN (LYSOZYME) | | Authors: | Dong, J, Boggon, T.J, Chayen, N.E, Raftery, J, Bi, R.C, Helliwell, J.R. | | Deposit date: | 1998-09-18 | | Release date: | 1998-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bound-solvent structures for microgravity-, ground control-, gel- and microbatch-grown hen egg-white lysozyme crystals at 1.8 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
3N12
 
 | | Crystal stricture of chitinase in complex with zinc atoms from Bacillus cereus NCTU2 | | Descriptor: | ACETIC ACID, Chitinase A, ZINC ION | | Authors: | Hsieh, Y.-C, Wu, Y.-J, Wu, W.-G, Li, Y.-K, Chen, C.-J. | | Deposit date: | 2010-05-15 | | Release date: | 2010-08-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structures of bacillus cereus NCTU2 chitinase complexes with chitooligomers reveal novel substrate binding for catalysis: a chitinase without chitin-binding and insertion domains
J.Biol.Chem., 285, 2010
|
|
8OYE
 
 | | Clostridium perfringens chitinase CP4_3455 E196Q with chitin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitodextrinase, DIMETHYL SULFOXIDE | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-05-04 | | Release date: | 2023-07-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Clostridium perfringens chitinases, key enzymes during early stages of necrotic enteritis in broiler chickens.
Plos Pathog., 20, 2024
|
|
1C3V
 
 | | DIHYDRODIPICOLINATE REDUCTASE FROM MYCOBACTERIUM TUBERCULOSIS COMPLEXED WITH NADPH AND PDC | | Descriptor: | DIHYDRODIPICOLINATE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PYRIDINE-2,6-DICARBOXYLIC ACID, ... | | Authors: | Cirilli, M, Zheng, R, Scapin, G, Blanchard, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 1999-07-28 | | Release date: | 2003-08-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | The three-dimensional structures of the Mycobacterium tuberculosis
dihydrodipicolinate reductase-NADH-2,6-PDC and -NADPH-2,6-PDC complexes.
Structural and mutagenic analysis of relaxed nucleotide specificity
Biochemistry, 42, 2003
|
|
8P5D
 
 | | Spraguea lophii ribosome in the closed conformation by cryo sub tomogram averaging | | Descriptor: | 40S Ribosomal protein S19, 40S ribosomal protein S0, 40S ribosomal protein S10, ... | | Authors: | Gil Diez, P, McLaren, M, Isupov, M.N, Daum, B, Conners, R, Williams, B. | | Deposit date: | 2023-05-23 | | Release date: | 2023-06-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (10.8 Å) | | Cite: | CryoEM reveals that ribosomes in microsporidian spores are locked in a dimeric hibernating state.
Nat Microbiol, 8, 2023
|
|
5IY8
 
 | | Human holo-PIC in the initial transcribing state | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB10, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | He, Y, Yan, C, Fang, J, Inouye, C, Tjian, R, Ivanov, I, Nogales, E. | | Deposit date: | 2016-03-24 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Near-atomic resolution visualization of human transcription promoter opening.
Nature, 533, 2016
|
|
9JDT
 
 | |
1T25
 
 | | Plasmodium falciparum lactate dehydrogenase complexed with NADH and 3-hydroxyisoxazole-4-carboxylic acid | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3-HYDROXYISOXAZOLE-4-CARBOXYLIC ACID, GLYCEROL, ... | | Authors: | Cameron, A, Read, J, Tranter, R, Winter, V.J, Sessions, R.B, Brady, R.L, Vivas, L, Easton, A, Kendrick, H, Croft, S.L, Barros, D, Lavandera, J.L, Martin, J.J, Risco, F, Garcia-Ochoa, S, Gamo, F.J, Sanz, L, Leon, L, Ruiz, J.R, Gabarro, R, Mallo, A, De Las Heras, F.G. | | Deposit date: | 2004-04-20 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification and Activity of a Series of Azole-based Compounds with Lactate Dehydrogenase-directed Anti-malarial Activity.
J.Biol.Chem., 279, 2004
|
|
5IMP
 
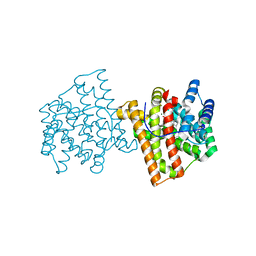 | | Crystal structure of N299A Aspergillus terreus aristolochene synthase complexed with (1S,8S,9aR)-1,9a-dimethyl-8-(prop-1-en-2-yl)decahydroquinolizin-5-ium | | Descriptor: | (1S,5S,8S,9aR)-1,9a-dimethyl-8-(prop-1-en-2-yl)octahydro-2H-quinolizinium, Aristolochene synthase, GLYCEROL, ... | | Authors: | Chen, M, Christianson, D.W. | | Deposit date: | 2016-03-06 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.038 Å) | | Cite: | Probing the Role of Active Site Water in the Sesquiterpene Cyclization Reaction Catalyzed by Aristolochene Synthase.
Biochemistry, 55, 2016
|
|
8PFV
 
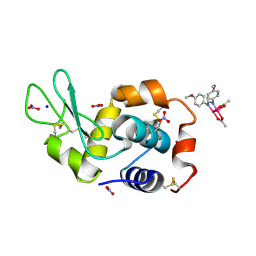 | | X-ray structure of the adduct formed upon reaction of Lysozyme with [Ru2Cl(DAniF)(O2CCH3)3] in condition A | | Descriptor: | 9,11-bis(4-methoxyphenyl)-3,7-dimethyl-2,4,6,8-tetraoxa-9,11-diaza-1$l^{4},5$l^{4}-diruthenatricyclo[3.3.3.0^{1,5}]undecane, CHLORIDE ION, Lysozyme C, ... | | Authors: | Teran, A, Ferraro, G, Merlino, A. | | Deposit date: | 2023-06-16 | | Release date: | 2023-07-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Charge effect in protein metalation reactions by diruthenium complexes
Inorg Chem Front, 2023
|
|
5NIL
 
 | | Structure of the MacAB-TolC ABC-type tripartite multidrug efflux pump-MacB section | | Descriptor: | Macrolide export ATP-binding/permease protein MacB, Macrolide export protein MacA, Outer membrane protein TolC | | Authors: | Fitzpatrick, A.W.P, Llabres, S, Neuberger, A, Blaza, J.N, Bai, X.-C, Okada, U, Murakami, S, van Veen, H.W, Zachariae, U, Scheres, S.H.W, Luisi, B.F, Du, D. | | Deposit date: | 2017-03-24 | | Release date: | 2017-05-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Structure of the MacAB-TolC ABC-type tripartite multidrug efflux pump.
Nat Microbiol, 2, 2017
|
|
5IOU
 
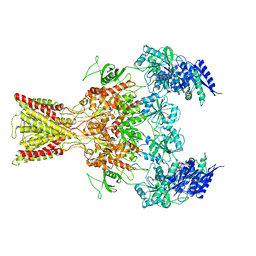 | | Cryo-EM structure of GluN1/GluN2B NMDA receptor in the glutamate/glycine-bound conformation | | Descriptor: | GLUTAMIC ACID, GLYCINE, Ionotropic glutamate receptor subunit NR2B, ... | | Authors: | Zhu, S, Stein, A.R, Yoshioka, C, Lee, C.H, Goehring, A, Mchaourab, S.H, Gouaux, E. | | Deposit date: | 2016-03-09 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Mechanism of NMDA Receptor Inhibition and Activation.
Cell, 165, 2016
|
|
5IPU
 
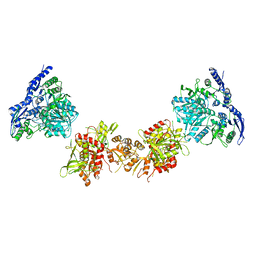 | | Cryo-EM structure of GluN1/GluN2B NMDA receptor in the DCKA/D-APV-bound conformation, state 6 | | Descriptor: | Ionotropic glutamate receptor subunit NR2B, N-methyl-D-aspartate receptor subunit NR1-8a | | Authors: | Zhu, S, Stein, A.R, Yoshioka, C, Lee, C.H, Goehring, A, Mchaourab, S.H, Gouaux, E. | | Deposit date: | 2016-03-09 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (15.4 Å) | | Cite: | Mechanism of NMDA Receptor Inhibition and Activation.
Cell, 165, 2016
|
|
8PFY
 
 | |
5ND2
 
 | | Microtubule-bound MKLP2 motor domain in the presence of ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Atherton, J, Yu, I.M, Cook, A, Muretta, J.M, Joseph, A.P, Major, J, Sourigues, Y, Clause, J, Topf, M, Rosenfeld, S.S, Houdusse, A, Moores, C.A. | | Deposit date: | 2017-03-07 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | The divergent mitotic kinesin MKLP2 exhibits atypical structure and mechanochemistry.
Elife, 6, 2017
|
|
8PFU
 
 | | X-ray structure of the adduct formed upon reaction of Lysozyme with K3[Ru2(CO3)4] in condition A | | Descriptor: | 6,8-bis(4-fluorophenyl)-1,5-bis(oxidanyl)-2,4-dioxa-6,8-diaza-1$l^{4},5$l^{4}-diruthenabicyclo[3.3.0]octan-3-one, CARBONATE ION, Lysozyme C, ... | | Authors: | Teran, A, Ferraro, G, Merlino, A. | | Deposit date: | 2023-06-16 | | Release date: | 2023-07-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Charge effect in protein metalation reactions by diruthenium complexes
Inorg Chem Front, 2023
|
|
8PFT
 
 | | X-ray structure of the adduct formed upon reaction of Lysozyme with K2[Ru2(D-p-FPhF)(CO3)3] in condition A | | Descriptor: | 6,8-bis(4-fluorophenyl)-1,5-bis(oxidanyl)-2,4-dioxa-6,8-diaza-1$l^{4},5$l^{4}-diruthenabicyclo[3.3.0]octan-3-one, CHLORIDE ION, Lysozyme C, ... | | Authors: | Teran, A, Ferraro, G, Merlino, A. | | Deposit date: | 2023-06-16 | | Release date: | 2023-07-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.304 Å) | | Cite: | Charge effect in protein metalation reactions by diruthenium complexes
Inorg Chem Front, 2023
|
|
1TDH
 
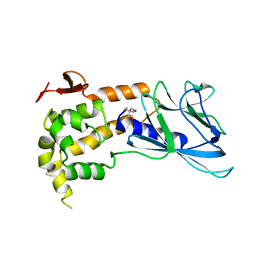 | | Crystal structure of human endonuclease VIII-like 1 (NEIL1) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, nei endonuclease VIII-like 1 | | Authors: | Doublie, S, Bandaru, V, Bond, J.P, Wallace, S.S. | | Deposit date: | 2004-05-22 | | Release date: | 2004-07-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of human endonuclease VIII-like 1 (NEIL1) reveals a zincless finger motif required for glycosylase activity.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
8IR7
 
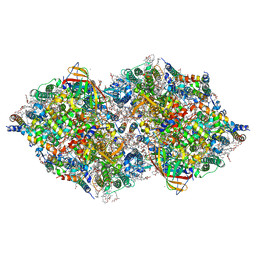 | | XFEL structure of cyanobacterial photosystem II following one flash (1F) with a 200-nanosecond delay | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, H, Suga, M, Shen, J.R. | | Deposit date: | 2023-03-17 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Oxygen-evolving photosystem II structures during S 1 -S 2 -S 3 transitions.
Nature, 626, 2024
|
|
5IR5
 
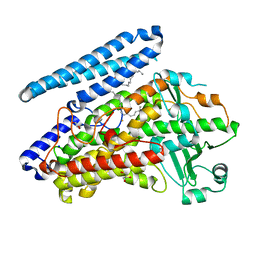 | | Crystal structure of wild-type bacterial lipoxygenase from Pseudomonas aeruginosa PA-LOX with space group P21212 at 1.9 A resolution | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradec-5-enoyloxy)propyl (11Z)-octadec-11-enoate, Arachidonate 15-lipoxygenase, FE (II) ION, ... | | Authors: | Kalms, J, Banthiya, S, Galemou Yoga, E, Kuhn, H, Scheerer, P. | | Deposit date: | 2016-03-12 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional basis of phospholipid oxygenase activity of bacterial lipoxygenase from Pseudomonas aeruginosa.
Biochim.Biophys.Acta, 1861, 2016
|
|
