5ZKC
 
 | | Crystal structure of rationally thermostabilized M2 muscarinic acetylcholine receptor bound with NMS | | Descriptor: | Muscarinic acetylcholine receptor M2,Apo-cytochrome b562,Muscarinic acetylcholine receptor M2, N-methyl scopolamine | | Authors: | Suno, R, Maeda, S, Yasuda, S, Yamashita, K, Hirata, K, Horita, S, Tawaramoto, M.S, Tsujimoto, H, Murata, T, Kinoshita, M, Yamamoto, M, Kobilka, B.K, Iwata, S, Kobayashi, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-11-21 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor
Nat. Chem. Biol., 14, 2018
|
|
6C51
 
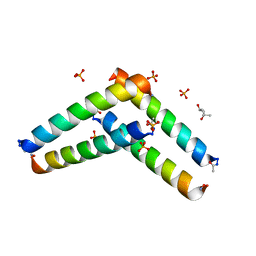 | | Cross-alpha Amyloid-like Structure alphaAmL | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cross-alpha Amyloid-like Structure alphaAmL, PHOSPHATE ION | | Authors: | Liu, L, Zhang, S.Q. | | Deposit date: | 2018-01-13 | | Release date: | 2018-08-15 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Designed peptides that assemble into cross-alpha amyloid-like structures.
Nat. Chem. Biol., 14, 2018
|
|
2Q1J
 
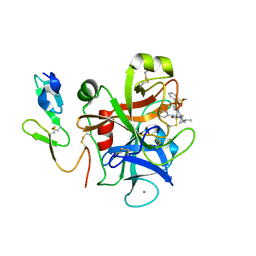 | | The discovery of glycine and related amino acid-based factor xa inhibitors | | Descriptor: | 1-(butyl{[(4-chlorophenyl)amino]carbonyl}amino)-N-[3-fluoro-2'-(methylsulfonyl)biphenyl-4-yl]cyclopropanecarboxamide, Activated factor Xa heavy chain (EC 3.4.21.6), CALCIUM ION, ... | | Authors: | Kohrt, J.T, Filipski, K.J, Cody, W.L, Bigge, C.F, Zhang, E, Finzel, B.C. | | Deposit date: | 2007-05-24 | | Release date: | 2007-08-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Discovery of Glycine and Related Amino Acid-Based Factor Xa Inhibitors
BIOORG.MED.CHEM., 14, 2006
|
|
6BPU
 
 | | Crystal structure of ferrous form of the F2-Tyr157 human cysteine dioxygenase with both uncrosslinked and crosslinked cofactor | | Descriptor: | Cysteine dioxygenase type 1, FE (II) ION, GLYCEROL, ... | | Authors: | Liu, A, Li, J, Shin, I. | | Deposit date: | 2017-11-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cleavage of a carbon-fluorine bond by an engineered cysteine dioxygenase.
Nat. Chem. Biol., 14, 2018
|
|
6BLH
 
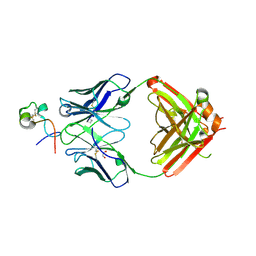 | | RSV G central conserved region bound to Fab CB017.5 | | Descriptor: | 1,2-ETHANEDIOL, Fab CB017.5 heavy chain, Fab CB017.5 light chain, ... | | Authors: | Jones, H.G, McLellan, J.S, Langedijk, J.P. | | Deposit date: | 2017-11-10 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for recognition of the central conserved region of RSV G by neutralizing human antibodies.
PLoS Pathog., 14, 2018
|
|
6BUR
 
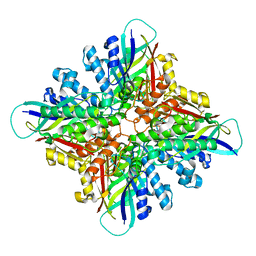 | |
6C0G
 
 | |
2RKL
 
 | | Crystal Structure of S.cerevisiae Vta1 C-terminal domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Vacuolar protein sorting-associated protein VTA1 | | Authors: | Xiao, J, Xia, H, Zhou, J, Xu, Z. | | Deposit date: | 2007-10-16 | | Release date: | 2008-01-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of vta1 function in the multivesicular body sorting pathway.
Dev.Cell, 14, 2008
|
|
6C4Y
 
 | | Cross-alpha Amyloid-like Structure alphaAmG | | Descriptor: | Cross-alpha Amyloid-like Structure alphaAmG | | Authors: | Liu, L, Zhang, S.Q. | | Deposit date: | 2018-01-13 | | Release date: | 2018-08-15 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Designed peptides that assemble into cross-alpha amyloid-like structures.
Nat. Chem. Biol., 14, 2018
|
|
6BUM
 
 | | Crystal structures of cyanuric acid hydrolase from Moorella thermoacetica | | Descriptor: | 1,3-PROPANDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Shi, K, Cho, S, Seffernick, J.L, Bera, A, Wackett, L.P, Aihara, H. | | Deposit date: | 2017-12-11 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structures of Moorella thermoacetica cyanuric acid hydrolase reveal conformational flexibility and asymmetry important for catalysis.
Plos One, 14, 2019
|
|
2OKJ
 
 | | The X-ray crystal structure of the 67kDa isoform of Glutamic Acid Decarboxylase (GAD67) | | Descriptor: | 4-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]BUTANOIC ACID, GAMMA-AMINO-BUTANOIC ACID, Glutamate decarboxylase 1 | | Authors: | Buckle, A.M, Fenalti, G, Law, R.H.P, Whisstock, J.C. | | Deposit date: | 2007-01-17 | | Release date: | 2007-03-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | GABA production by glutamic acid decarboxylase is regulated by a dynamic catalytic loop.
Nat.Struct.Mol.Biol., 14, 2007
|
|
6CDG
 
 | | GID4 fragment in complex with a peptide | | Descriptor: | Glucose-induced degradation protein 4 homolog, Hexapeptide PGLWKS, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-08 | | Release date: | 2018-03-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis of GID4-mediated recognition of degrons for the Pro/N-end rule pathway.
Nat. Chem. Biol., 14, 2018
|
|
5ZK3
 
 | | Crystal structure of rationally thermostabilized M2 muscarinic acetylcholine receptor bound with QNB | | Descriptor: | (3R)-1-azabicyclo[2.2.2]oct-3-yl hydroxy(diphenyl)acetate, Muscarinic acetylcholine receptor M2,Apo-cytochrome b562,Muscarinic acetylcholine receptor M2 | | Authors: | Suno, R, Maeda, S, Yasuda, S, Yamashita, K, Hirata, K, Horita, S, Tawaramoto, M.S, Tsujimoto, H, Murata, T, Kinoshita, M, Yamamoto, M, Kobilka, B.K, Iwata, S, Kobayashi, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-11-21 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor
Nat. Chem. Biol., 14, 2018
|
|
6BGF
 
 | | Crystal structure of cysteine-bound ferrous form of the crosslinked human cysteine dioxygenase | | Descriptor: | CYSTEINE, Cysteine dioxygenase type 1, FE (II) ION, ... | | Authors: | Liu, A, Li, J, Shin, I. | | Deposit date: | 2017-10-28 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Cleavage of a carbon-fluorine bond by an engineered cysteine dioxygenase.
Nat.Chem.Biol., 14, 2018
|
|
6BYN
 
 | | Crystal structure of WDR5-Mb(S4) monobody complex | | Descriptor: | WD repeat-containing protein 5, WDR5-binding Monobody, Mb(S4) | | Authors: | Gupta, A, Koide, S. | | Deposit date: | 2017-12-21 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Facile target validation in an animal model with intracellularly expressed monobodies.
Nat. Chem. Biol., 14, 2018
|
|
6C0H
 
 | |
2Q2E
 
 | |
2Q6J
 
 | | Crystal Structure of Estrogen Receptor alpha Complexed to a B-N Substituted Ligand | | Descriptor: | 4-[(DIMESITYLBORYL)(2,2,2-TRIFLUOROETHYL)AMINO]PHENOL, Estrogen receptor, GRIP peptide | | Authors: | Zhou, H, Nettles, K.W, Bruning, J.B, Kim, Y, Joachimiak, A, Sharma, S, Carlson, K.E, Stossi, F, Katzenellenbogen, B.S, Greene, G.L, Katzenellenbogen, J.A. | | Deposit date: | 2007-06-05 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Elemental isomerism: a boron-nitrogen surrogate for a carbon-carbon double bond increases the chemical diversity of estrogen receptor ligands
Chem.Biol., 14, 2007
|
|
6BN9
 
 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET70 PROTAC | | Descriptor: | Bromodomain-containing protein 4, DNA damage-binding protein 1,DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Nowak, R.P, DeAngelo, S.L, Buckley, D, Bradner, J.E, Fischer, E.S. | | Deposit date: | 2017-11-16 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.382 Å) | | Cite: | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
6BNB
 
 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET57 PROTAC | | Descriptor: | Bromodomain-containing protein 4, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Nowak, R.P, DeAngelo, S.L, Buckley, D, Ishoey, M, He, Z, Zhang, T, Bradner, J.E, Fischer, E.S. | | Deposit date: | 2017-11-16 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (6.343 Å) | | Cite: | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
6CD8
 
 | | Complex of GID4 fragment with short peptide | | Descriptor: | Glucose-induced degradation protein 4 homolog, Tetrapeptide PSRV, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-08 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis of GID4-mediated recognition of degrons for the Pro/N-end rule pathway.
Nat. Chem. Biol., 14, 2018
|
|
6CDC
 
 | | GID4 in complex with a tetrapeptide | | Descriptor: | Glucose-induced degradation protein 4 homolog, Tetrapeptide PGLW, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-08 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular basis of GID4-mediated recognition of degrons for the Pro/N-end rule pathway.
Nat. Chem. Biol., 14, 2018
|
|
6CDN
 
 | | Crystal structure of cysteine-bound ferrous form of the crosslinked Cl-Tyr157 human cysteine dioxygenase | | Descriptor: | CYSTEINE, Cysteine dioxygenase type 1, FE (II) ION, ... | | Authors: | Liu, A, Li, J, Shin, I. | | Deposit date: | 2018-02-08 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.055 Å) | | Cite: | Cleavage of a carbon-fluorine bond by an engineered cysteine dioxygenase.
Nat. Chem. Biol., 14, 2018
|
|
7KII
 
 | | Muscovy duck circovirus Rep domain complexed with a single-stranded DNA 10-mer comprising the cleavage site and Mn2+ | | Descriptor: | ATP-dependent helicase Rep, CALCIUM ION, DNA (5'-D(*TP*AP*TP*TP*AP*TP*TP*AP*CP*C)-3'), ... | | Authors: | Tompkins, K.J, Gordon, W.R, Shi, K. | | Deposit date: | 2020-10-23 | | Release date: | 2021-11-17 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Watson-Crick Base-Pairing Requirements for ssDNA Recognition and Processing in Replication-Initiating HUH Endonucleases.
Mbio, 14, 2023
|
|
7KIJ
 
 | | Muscovy duck circovirus Rep domain complexed with a single-stranded DNA 10-mer comprising the cleavage site | | Descriptor: | ATP-dependent helicase Rep, DNA (5'-D(*TP*AP*TP*TP*AP*TP*TP*AP*CP*C)-3'), GLYCEROL, ... | | Authors: | Tompkins, K.J, Gordon, W.R, Shi, K. | | Deposit date: | 2020-10-23 | | Release date: | 2021-11-17 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Watson-Crick Base-Pairing Requirements for ssDNA Recognition and Processing in Replication-Initiating HUH Endonucleases.
Mbio, 14, 2023
|
|
