6WVO
 
 | | Crystal Structure of Recombinant Human Acetylcholinesterase In Complex with GD and HI-6 | | Descriptor: | (1R)-1,2,2-TRIMETHYLPROPYL (S)-METHYLPHOSPHINATE, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | McGuire, J.R, Bester, S.M, Pegan, S.D, Height, J.J. | | Deposit date: | 2020-05-06 | | Release date: | 2021-03-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural and Biochemical Insights into the Inhibition of Human Acetylcholinesterase by G-Series Nerve Agents and Subsequent Reactivation by HI-6.
Chem.Res.Toxicol., 34, 2021
|
|
6Q96
 
 | |
7T1J
 
 | | Crystal structure of RUBISCO from Rhodospirillaceae bacterium BRH_c57 | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Pereira, J.H, Liu, A.K, Shih, P.M, Adams, P.D. | | Deposit date: | 2021-12-02 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural plasticity enables evolution and innovation of RuBisCO assemblies.
Sci Adv, 8, 2022
|
|
9HL5
 
 | | Crystal structure of halo-tolerant PETase from marine metagenome (HaloPETase1) | | Descriptor: | 1,2-ETHANEDIOL, Alpha/beta hydrolase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Turak, O, Kriegel, M, Hocker, B. | | Deposit date: | 2024-12-04 | | Release date: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | A third type of PETase from the marine Halopseudomonas lineage.
Protein Sci., 34, 2025
|
|
7SUG
 
 | | Structure of CHK1 10-pt. mutant complex with LRRK2 inhibitor 09 | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-{[5-methyl-1-(oxan-4-yl)-1H-pyrazol-4-yl]amino}quinazolin-8-yl)cyclopropane-1-carbonitrile, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L. | | Deposit date: | 2021-11-17 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structure-Guided Discovery of Aminoquinazolines as Brain-Penetrant and Selective LRRK2 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7SUH
 
 | | Structure of CHK1 10-pt. mutant complex with LRRK2 inhibitor 15 | | Descriptor: | 1-[5-chloro-4-({6-chloro-7-[1-(oxetan-3-yl)piperidin-4-yl]quinazolin-2-yl}amino)-1H-pyrazol-1-yl]-2-methylpropan-2-ol, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L. | | Deposit date: | 2021-11-17 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structure-Guided Discovery of Aminoquinazolines as Brain-Penetrant and Selective LRRK2 Inhibitors.
J.Med.Chem., 65, 2022
|
|
3ILK
 
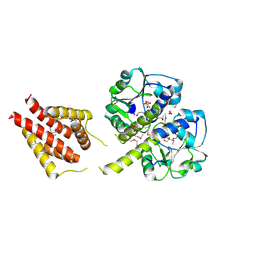 | | The structure of a probable methylase family protein from Haemophilus influenzae Rd KW20 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, SULFATE ION, ... | | Authors: | Tan, K, Li, H, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-08-07 | | Release date: | 2009-09-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The structure of a probable methylase family protein from Haemophilus influenzae Rd KW20
To be Published
|
|
8J1N
 
 | | Structure of human UCP1 in the DNP-bound state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2,4-DINITROPHENOL, CARDIOLIPIN, ... | | Authors: | Chen, L, Kang, Y. | | Deposit date: | 2023-04-13 | | Release date: | 2023-06-21 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Structural basis for the binding of DNP and purine nucleotides onto UCP1.
Nature, 620, 2023
|
|
6ETD
 
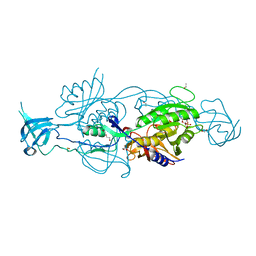 | |
6PTX
 
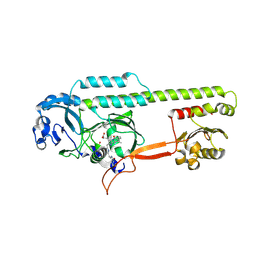 | | Dark, 100K, PCM Myxobacterial Phytochrome, P2, Wild Type, | | Descriptor: | 3-[(2Z)-2-({3-(2-carboxyethyl)-5-[(E)-(4-ethenyl-3-methyl-5-oxo-1,5-dihydro-2H-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-2-yl}methylidene)-5-{(Z)-[(3E,4S)-3-ethylidene-4-methyl-5-oxopyrrolidin-2-ylidene]methyl}-4-methyl-2H-pyrrol-3-yl]propanoic acid, Photoreceptor-histidine kinase BphP | | Authors: | Pandey, S, Schmidt, M, Stojkovic, E.A. | | Deposit date: | 2019-07-16 | | Release date: | 2019-10-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | High-resolution crystal structures of a myxobacterial phytochrome at cryo and room temperatures.
Struct Dyn., 6, 2019
|
|
5OAE
 
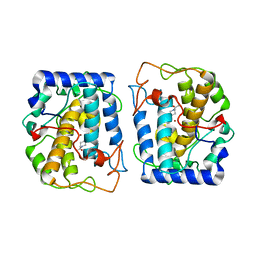 | | Crystal Structure of tyrosinase from Bacillus megaterium with SVF inhibitor in the active site | | Descriptor: | 1-[4-[(4-fluorophenyl)methyl]piperidin-1-yl]ethanone, COPPER (II) ION, Tyrosinase | | Authors: | Deri, B, Gitto, R, Pazy Benhar, Y, Fishman, A. | | Deposit date: | 2017-06-21 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Targeting Tyrosinase: Development and Structural Insights of Novel Inhibitors Bearing Arylpiperidine and Arylpiperazine Fragments.
J. Med. Chem., 61, 2018
|
|
3ABN
 
 | | Crystal structure of (Pro-Pro-Gly)4-Hyp-Asp-Gly-(Pro-Pro-Gly)4 at 1.02 A | | Descriptor: | collagen-like peptide | | Authors: | Okuyama, K, Shimura, M, Kawaguchi, T, Noguchi, K, Mizuno, K, Bachinger, H.P. | | Deposit date: | 2009-12-16 | | Release date: | 2010-12-01 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Crystal structure of the collagen model peptide (Pro-Pro-Gly)4 -Hyp-Asp-Gly-(Pro-Pro-Gly)4 at 1.0 angstrom resolution.
Biopolymers, 99, 2013
|
|
7T1G
 
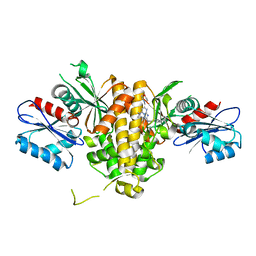 | | Crystal structure of CAB1 Pantothenate Kinase from Saccharomyces cerevisiae in complex with compound YU385595 | | Descriptor: | (8S)-N~2~-[(4-tert-butylphenyl)methyl]-N~7~,N~7~-dimethyl-5-[(morpholin-4-yl)methyl][1,2,4]triazolo[1,5-a]pyrimidine-2,7-diamine, Pantothenate kinase CAB1 | | Authors: | Gihaz, S, Ben Mamoun, C. | | Deposit date: | 2021-12-01 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution crystal structure and chemical screening reveal pantothenate kinase as a new target for antifungal development.
Structure, 30, 2022
|
|
6R1N
 
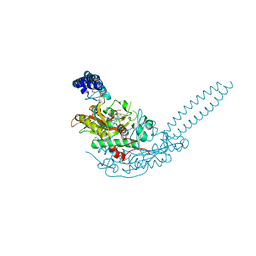 | | Crystal structure of S. aureus seryl-tRNA synthetase complexed to seryl sulfamoyl adenosine | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Salimraj, R, Cain, R, Roper, D.I. | | Deposit date: | 2019-03-14 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure-Guided Enhancement of Selectivity of Chemical Probe Inhibitors Targeting Bacterial Seryl-tRNA Synthetase.
J.Med.Chem., 62, 2019
|
|
7SE8
 
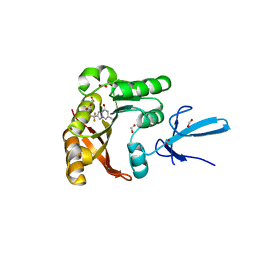 | | Crystal structure of human Fibrillarin in complex with fragment 1 from cocktail soak | | Descriptor: | (5S)-3-methyl-7-(trifluoromethyl)pyrrolo[1,2-a]pyrazin-1(2H)-one, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Shi, Y, El-Deeb, I.M, Masic, V, Hartley-Tassell, L, Maggioni, A, von Itzstein, M, Ve, T. | | Deposit date: | 2021-09-30 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Cofactor Competitive Inhibitors against the Human Methyltransferase Fibrillarin.
Pharmaceuticals, 15, 2021
|
|
6R1M
 
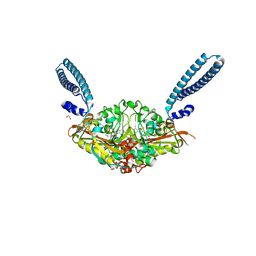 | | Crystal structure of E. coli seryl-tRNA synthetase complexed to seryl sulfamoyl adenosine | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, PHOSPHATE ION, ... | | Authors: | Salimraj, R, Cain, R, Roper, D.I. | | Deposit date: | 2019-03-14 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Guided Enhancement of Selectivity of Chemical Probe Inhibitors Targeting Bacterial Seryl-tRNA Synthetase.
J.Med.Chem., 62, 2019
|
|
7SE9
 
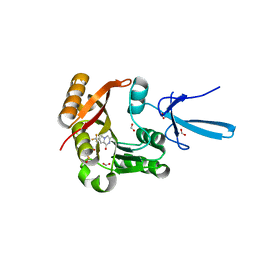 | | Crystal structure of human Fibrillarin in complex with compound 1 from single soak | | Descriptor: | (5S)-3-methyl-7-(trifluoromethyl)pyrrolo[1,2-a]pyrazin-1(2H)-one, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Shi, Y, El-Deeb, I.M, Masic, V, Hartley-Tassell, L, Maggioni, A, von Itzstein, M, Ve, T. | | Deposit date: | 2021-09-30 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Cofactor Competitive Inhibitors against the Human Methyltransferase Fibrillarin.
Pharmaceuticals, 15, 2021
|
|
5A6W
 
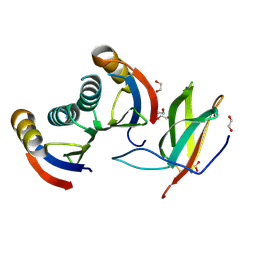 | | Complex of rice blast (Magnaporthe oryzae) effector protein AVR-PikD with the HMA domain of Pikp1 from rice (Oryza sativa) | | Descriptor: | 1,2-ETHANEDIOL, AVR-PIK PROTEIN, RESISTANCE PROTEIN PIKP-1, ... | | Authors: | Maqbool, A, Saitoh, H, Franceschetti, M, Stevenson, C.E, Uemura, A, Kanzaki, H, Kamoun, S, Terauchi, R, Banfield, M.J. | | Deposit date: | 2015-07-01 | | Release date: | 2015-08-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of pathogen recognition by an integrated HMA domain in a plant NLR immune receptor.
Elife, 4, 2015
|
|
5HNZ
 
 | | Structural basis of backwards motion in kinesin-14: plus-end directed nKn669 in the nucleotide-free state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Shigematsu, H, Yokoyama, T, Kikkawa, M, Shirouzu, M, Nitta, R. | | Deposit date: | 2016-01-19 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structural Basis of Backwards Motion in Kinesin-1-Kinesin-14 Chimera: Implication for Kinesin-14 Motility
Structure, 24, 2016
|
|
7S4K
 
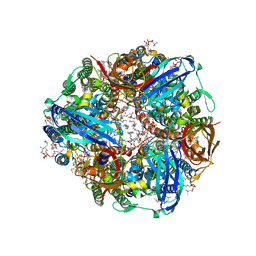 | | CryoEM structure of Methylococcus capsulatus (Bath) pMMO in a native lipid nanodisc at 2.34 Angstrom resolution | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Ammonia monooxygenase/methane monooxygenase, ... | | Authors: | Koo, C.W, Rosenzweig, A.C. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Recovery of particulate methane monooxygenase structure and activity in a lipid bilayer.
Science, 375, 2022
|
|
7S4I
 
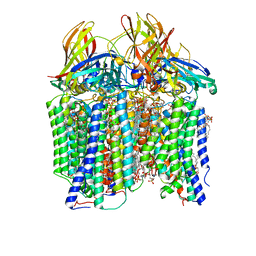 | | CryoEM structure of Methylococcus capsulatus (Bath) pMMO in a native lipid nanodisc at 2.26 Angstrom resolution | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Ammonia monooxygenase/methane monooxygenase, ... | | Authors: | Koo, C.W, Rosenzweig, A.C. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | Recovery of particulate methane monooxygenase structure and activity in a lipid bilayer.
Science, 375, 2022
|
|
6F4U
 
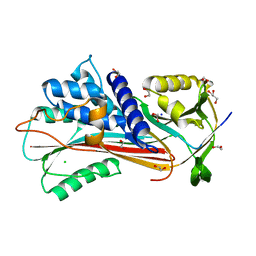 | |
7S4J
 
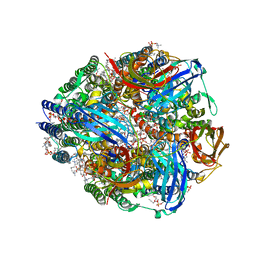 | | CryoEM structure of Methylococcus capsulatus (Bath) pMMO in a native lipid nanodisc at 2.16 Angstrom resolution | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Ammonia monooxygenase/methane monooxygenase, ... | | Authors: | Koo, C.W, Rosenzweig, A.C. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.16 Å) | | Cite: | Recovery of particulate methane monooxygenase structure and activity in a lipid bilayer.
Science, 375, 2022
|
|
7S4H
 
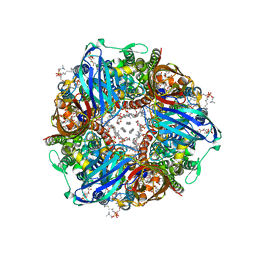 | | CryoEM structure of Methylococcus capsulatus (Bath) pMMO in a native lipid nanodisc at 2.14 Angstrom resolution | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Ammonia monooxygenase/methane monooxygenase, ... | | Authors: | Koo, C.W, Rosenzweig, A.C. | | Deposit date: | 2021-09-08 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.14 Å) | | Cite: | Recovery of particulate methane monooxygenase structure and activity in a lipid bilayer.
Science, 375, 2022
|
|
7CGP
 
 | | Cryo-EM structure of the human mitochondrial translocase TIM22 complex at 3.7 angstrom. | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Acylglycerol kinase, mitochondrial, ... | | Authors: | Qi, L, Wang, Q, Guan, Z, Yan, C, Yin, P. | | Deposit date: | 2020-07-01 | | Release date: | 2020-10-14 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the human mitochondrial translocase TIM22 complex.
Cell Res., 31, 2021
|
|
