1E19
 
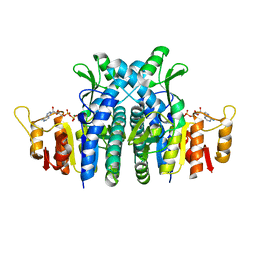 | | Structure of the carbamate kinase-like carbamoyl phosphate synthetase from the hyperthermophilic archaeon Pyrococcus furiosus bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CARBAMATE KINASE, MAGNESIUM ION | | Authors: | Ramon-Maiques, S, Marina, A, Uriarte, M, Fita, I, Rubio, V. | | Deposit date: | 2000-04-28 | | Release date: | 2000-07-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The 1.5-A Resolution Crystal Structure of the Carbamate Kinase-Like Carbamoyl Phosphate Synthetase from the Hyperthermophilic Archaeon Pyrococcus Furiosus, Bound to Adp, Confirms that This Thermoestable Enzyme is a Carbamate Kinase, and Provides Insights Into Substrate Binding and Stability in Carbamate Kinases
J.Mol.Biol., 299, 2000
|
|
1EQQ
 
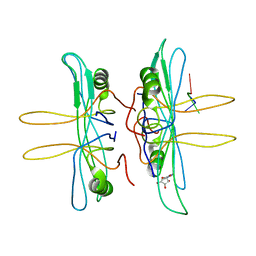 | | SINGLE STRANDED DNA BINDING PROTEIN AND SSDNA COMPLEX | | Descriptor: | 5'-R(*(5MU)P*(5MU)P*(5MU))-3', SINGLE STRANDED DNA BINDING PROTEIN | | Authors: | Matsumoto, T, Morimoto, Y, Shibata, N, Yasuoka, N, Shimamoto, N. | | Deposit date: | 2000-04-06 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Roles of functional loops and the C-terminal segment of a single-stranded DNA binding protein elucidated by X-Ray structure analysis
J.Biochem.(Tokyo), 127, 2000
|
|
2OSF
 
 | | Inhibition of Carbonic Anhydrase II by Thioxolone: A Mechanistic and Structural Study | | Descriptor: | 4-MERCAPTOBENZENE-1,3-DIOL, Carbonic anhydrase 2, S-(2,4-dihydroxyphenyl) hydrogen thiocarbonate, ... | | Authors: | Albert, A.B, Caroli, G, Govindasamy, L, Agbandje-McKenna, M, McKenna, R, Tripp, B.C. | | Deposit date: | 2007-02-05 | | Release date: | 2008-05-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Inhibition of carbonic anhydrase II by thioxolone: a mechanistic and structural study.
Biochemistry, 47, 2008
|
|
1EFG
 
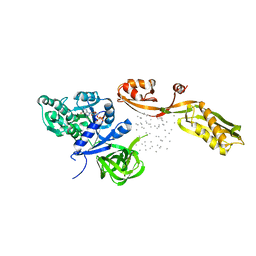 | | THE CRYSTAL STRUCTURE OF ELONGATION FACTOR G COMPLEXED WITH GDP, AT 2.7 ANGSTROMS RESOLUTION | | Descriptor: | ELONGATION FACTOR G, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Czworkowski, J, Wang, J, Steitz, T.A, Moore, P.B. | | Deposit date: | 1994-10-17 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of elongation factor G complexed with GDP, at 2.7 A resolution.
EMBO J., 13, 1994
|
|
1EAQ
 
 | | The RUNX1 Runt domain at 1.25A resolution: A structural switch and specifically bound chloride ions modulate DNA binding | | Descriptor: | CHLORIDE ION, RUNT-RELATED TRANSCRIPTION FACTOR 1 | | Authors: | Backstrom, S, Wolf-Watz, M, Grundstrom, C, Hard, T, Grundstrom, T, Sauer, U.H. | | Deposit date: | 2001-07-14 | | Release date: | 2002-09-12 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The Runx1 Runt Domain at 1.25 A Resolution: A Structural Switch and Specifically Bound Chloride Ions Modulate DNA Binding
J.Mol.Biol., 322, 2002
|
|
1EQ6
 
 | |
2BLV
 
 | | Trypsin before a high dose x-ray "burn" | | Descriptor: | BENZAMIDINE, CALCIUM ION, GLYCEROL, ... | | Authors: | Nanao, M.H, Ravelli, R.B. | | Deposit date: | 2005-03-08 | | Release date: | 2005-09-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Improving Radiation-Damage Substructures for Rip.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2BI0
 
 | | RV0216, A conserved hypothetical protein from Mycobacterium tuberculosis that is essential for bacterial survival during infection, has a double hotdogfold | | Descriptor: | CHLORIDE ION, HYPOTHETICAL PROTEIN RV0216 | | Authors: | Castell, A, Johansson, P, Unge, T, Jones, T.A, Backbro, K. | | Deposit date: | 2005-01-20 | | Release date: | 2005-04-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rv0216, a Conserved Hypothetical Protein from Mycobacterium Tuberculosis that is Essential for Bacterial Survival During Infection, Has a Double Hotdog Fold
Protein Sci., 14, 2005
|
|
2BVQ
 
 | | Structures of Three HIV-1 HLA-B5703-Peptide Complexes and Identification of Related HLAs Potentially Associated with Long-Term Non-Progression | | Descriptor: | BETA-2-MICROGLOBULIN, HIV-P24, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Stewart-Jones, G.B, Gillespie, G, Overton, I.M, Kaul, R, Roche, P, Mcmichael, A.J, Rowland-Jones, S, Jones, E.Y. | | Deposit date: | 2005-07-01 | | Release date: | 2005-09-07 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of three HIV-1 HLA-B*5703-peptide complexes and identification of related HLAs potentially associated with long-term nonprogression.
J Immunol., 175, 2005
|
|
1OUU
 
 | | CARBONMONOXY TROUT HEMOGLOBIN I | | Descriptor: | CARBON MONOXIDE, HEMOGLOBIN I, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tame, J, Wilson, J. | | Deposit date: | 1996-06-21 | | Release date: | 1997-01-11 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structures of trout Hb I in the deoxy and carbonmonoxy forms.
J.Mol.Biol., 259, 1996
|
|
1ORR
 
 | |
1OPB
 
 | |
1SN1
 
 | | STRUCTURE OF SCORPION NEUROTOXIN BMK M1 | | Descriptor: | PROTEIN (NEUROTOXIN BMK M1) | | Authors: | He, X.L, Li, H.M, Liu, X.Q, Zeng, Z.H, Wang, D.C. | | Deposit date: | 1998-11-12 | | Release date: | 1999-11-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of two alpha-like scorpion toxins: non-proline cis peptide bonds and implications for new binding site selectivity on the sodium channel.
J.Mol.Biol., 292, 1999
|
|
1SQ3
 
 | | Crystal structures of a novel open pore ferritin from the hyperthermophilic Archaeon Archaeoglobus fulgidus. | | Descriptor: | FE (III) ION, ferritin | | Authors: | Johnson, E, Cascio, D, Michael, S, Schroder, I. | | Deposit date: | 2004-03-17 | | Release date: | 2005-04-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of a tetrahedral open pore ferritin from the hyperthermophilic archaeon Archaeoglobus fulgidus.
Structure, 13, 2005
|
|
1P3N
 
 | | CORE REDESIGN BACK-REVERTANT I103V/CORE10 | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME, ... | | Authors: | Mooers, B.H, Datta, D, Baase, W.A, Zollars, E.S, Mayo, S.L, Matthews, B.W. | | Deposit date: | 2003-04-17 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Repacking the Core of T4 lysozyme by automated design
J.Mol.Biol., 332, 2003
|
|
1WKR
 
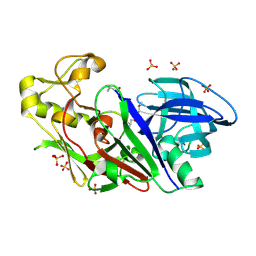 | | Crystal structure of aspartic proteinase from Irpex lacteus | | Descriptor: | Polyporopepsin, SULFATE ION, pepstatin | | Authors: | Fujimoto, Z, Fujii, Y, Kaneko, S, Kobayashi, H, Mizuno, H. | | Deposit date: | 2004-06-02 | | Release date: | 2004-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of Aspartic Proteinase from Irpex lacteus in Complex with Inhibitor Pepstatin
J.Mol.Biol., 341, 2004
|
|
1ORJ
 
 | | FLAGELLAR EXPORT CHAPERONE | | Descriptor: | flagellar protein FliS | | Authors: | Evdokimov, A.G, Phan, J, Tropea, J.E, Routzahn, K.M, Peters III, H.K, Pokross, M, Waugh, D.S. | | Deposit date: | 2003-03-13 | | Release date: | 2003-09-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Similar modes of polypeptide recognition by export chaperones in flagellar biosynthesis and type III secretion
Nat.Struct.Biol., 10, 2003
|
|
1ORW
 
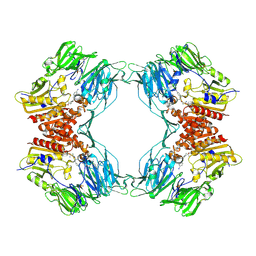 | | Crystal Structure of Porcine Dipeptidyl Peptidase IV (CD26) in Complex with a Peptidomimetic Inhibitor | | Descriptor: | (2S)-PYRROLIDIN-2-YLMETHYLAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Engel, M, Hoffmann, T, Wagner, L, Wermann, M, Heiser, U, Kiefersauer, R, Huber, R, Bode, W, Demuth, H.U, Brandstetter, H. | | Deposit date: | 2003-03-16 | | Release date: | 2003-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | The Crystal Structure of Dipeptidyl Peptidase IV (CD26) Reveals its Functional Regulation and Enzymatic Mechanism
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1P4I
 
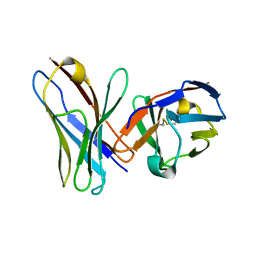 | | Crystal Structure of scFv against peptide GCN4 | | Descriptor: | ANTIBODY VARIABLE LIGHT CHAIN, antibody variable heavy chain | | Authors: | Zahnd, C, Spinelli, S, Luginbuhl, B, Jermutus, L, Amstutz, P, Cambillau, C, Pluckthun, A. | | Deposit date: | 2003-04-23 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Directed in Vitro Evolution and Crystallographic Analysis of a Peptide-binding Single Chain Antibody Fragment (scFv) with Low Picomolar Affinity.
J.Biol.Chem., 279, 2004
|
|
1P9G
 
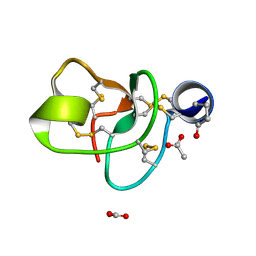 | | Crystal structure of a novel antifungal protein distinct with five disulfide bridges from Ecommia ulmoides Oliver at atomic resolution | | Descriptor: | ACETATE ION, EAFP 2 | | Authors: | Xiang, Y, Huang, R.H, Liu, X.Z, Wang, D.C. | | Deposit date: | 2003-05-12 | | Release date: | 2004-06-01 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (0.84 Å) | | Cite: | Crystal structure of a novel antifungal protein distinct with five disulfide bridges from Eucommia ulmoides Oliver at an atomic resolution.
J.Struct.Biol., 148, 2004
|
|
1OX8
 
 | | Crystal structure of SspB | | Descriptor: | Stringent starvation protein B | | Authors: | Song, H.K, Eck, M.J. | | Deposit date: | 2003-04-01 | | Release date: | 2003-08-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of degradation signal recognition by SspB, a specificity-enhancing factor for the ClpXP proteolytic machine
Mol.Cell, 12, 2003
|
|
2YAD
 
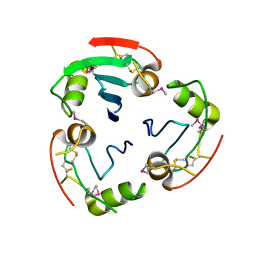 | | BRICHOS domain of Surfactant protein C precursor protein | | Descriptor: | SURFACTANT PROTEIN C BRICHOS DOMAIN | | Authors: | Askarieh, G, Siponen, M.I, Willander, H, Landreh, M, Westermark, P, Nordling, K, Keranen, H, Hermansson, E, Hamvas, A, Nogee, L.M, Bergman, T, Saenz, A, Casals, C, Aqvist, J, Jornvall, H, Presto, J, Johansson, J, Arrowsmith, C.H, Bountra, C, Collins, R, Edwards, A.M, Ekblad, T, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kol, S, Kotenyova, T, Kouznetsova, E, Moche, M, Nyman, T, Nordlund, P, Persson, C, Schuler, H, Thorsell, A.G, Tresaugues, L, van den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Berglund, H, Knight, S.D. | | Deposit date: | 2011-02-18 | | Release date: | 2012-02-15 | | Last modified: | 2014-08-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High Resolution Structure of a Bricos Domain and its Implications for Anti-Amyloid Chaperone Activity on Lung Surgactant Protein C.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1IIW
 
 | |
1SJT
 
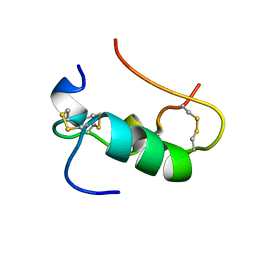 | | MINI-PROINSULIN, TWO CHAIN INSULIN ANALOG MUTANT: DES B30, HIS(B 10)ASP, PRO(B 28)ASP, NMR, 20 STRUCTURES | | Descriptor: | PROINSULIN | | Authors: | Hua, Q.X, Hu, S.Q, Jia, W.H, Chu, Y.C, Burke, G.T, Wang, S.H, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 1997-10-09 | | Release date: | 1998-03-18 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Mini-proinsulin and mini-IGF-I: homologous protein sequences encoding non-homologous structures.
J.Mol.Biol., 277, 1998
|
|
3E4B
 
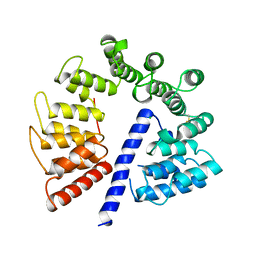 | | Crystal structure of AlgK from Pseudomonas fluorescens WCS374r | | Descriptor: | AlgK, CHLORIDE ION, GLYCEROL | | Authors: | Keiski, C.-L, Harwich, M, Jain, S, Neculai, A.M, Yip, P, Robinson, H, Whitney, J.C, Burrows, L.L, Ohman, D.E, Howell, P.L. | | Deposit date: | 2008-08-11 | | Release date: | 2009-08-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | AlgK is a TPR-containing protein and the periplasmic component of a novel exopolysaccharide secretin.
Structure, 18, 2010
|
|
