8WFL
 
 | |
8UFK
 
 | |
7BD9
 
 | | Notum Fragment 886 | | Descriptor: | (4-chloranyl-2-methyl-pyrazol-3-yl)-piperidin-1-yl-methanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-12-21 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
8IYT
 
 | | Crystal Structure of Serine Palmitoyltransferase complexed with D-methylserine | | Descriptor: | (2~{R})-2-methyl-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]-3-oxidanyl-propanoic acid, 1,2-ETHANEDIOL, Serine palmitoyltransferase | | Authors: | Takahashi, A, Murakami, T, Katayama, A, Miyahara, I, Kamiya, N, Ikushiro, H, Yano, T. | | Deposit date: | 2023-04-06 | | Release date: | 2024-04-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Racemization of the substrate and product by serine palmitoyltransferase from Sphingobacterium multivorum yields two enantiomers of the product from d-serine.
J.Biol.Chem., 300, 2024
|
|
7ASH
 
 | |
6XHL
 
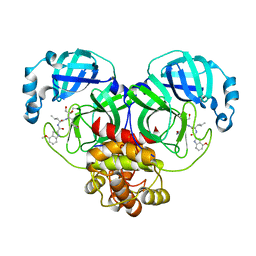 | | Covalent complex of SARS-CoV main protease with N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Descriptor: | 3C-like proteinase, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Gajiwala, K.S, Ferre, R.A, Ryan, K, Stewart, A.E. | | Deposit date: | 2020-06-19 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.471 Å) | | Cite: | Discovery of Ketone-Based Covalent Inhibitors of Coronavirus 3CL Proteases for the Potential Therapeutic Treatment of COVID-19.
J.Med.Chem., 63, 2020
|
|
4Y1V
 
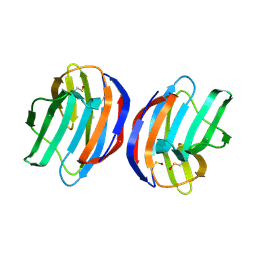 | | Complex of human Galectin-1 and Galbeta1-3GlcNAc | | Descriptor: | Galectin-1, beta-D-galactopyranose-(1-3)-methyl 2-acetamido-2-deoxy-beta-D-glucopyranoside | | Authors: | Lin, H.Y, Hsieh, T.J, Lin, C.H. | | Deposit date: | 2015-02-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural basis of human galectin-1 inhibition with Ki values in the micro- to nanomolar range
To Be Published
|
|
5KTB
 
 | |
7UBY
 
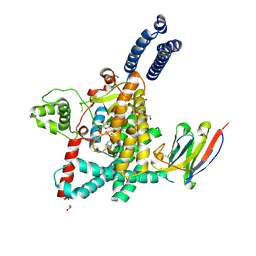 | | Structure of the GTD domain of Clostridium difficile toxin A in complex with VHH AH3 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Glucosyltransferase TcdA, ... | | Authors: | Chen, B, Rongsheng, J, Kay, P. | | Deposit date: | 2022-03-15 | | Release date: | 2022-11-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Neutralizing epitopes on Clostridioides difficile toxin A revealed by the structures of two camelid VHH antibodies.
Front Immunol, 13, 2022
|
|
8RYS
 
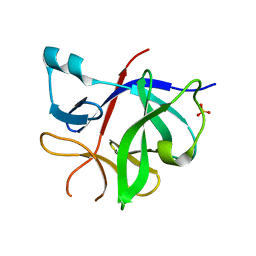 | | Human IL-1beta, unliganded | | Descriptor: | Interleukin-1 beta, SULFATE ION | | Authors: | Rondeau, J.-M, Lehmann, S. | | Deposit date: | 2024-02-09 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Ligandability Assessment of IL-1 beta by Integrated Hit Identification Approaches.
J.Med.Chem., 67, 2024
|
|
6RLN
 
 | | Crystal structure of RIP1 kinase in complex with GSK3145095 | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 1, ~{N}-[(3~{S})-7,9-bis(fluoranyl)-2-oxidanylidene-1,3,4,5-tetrahydro-1-benzazepin-3-yl]-3-(phenylmethyl)-1~{H}-1,2,4-triazole-5-carboxamide | | Authors: | Thorpe, J.H, Harris, P.A. | | Deposit date: | 2019-05-02 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Identification of a RIP1 Kinase Inhibitor Clinical Candidate (GSK3145095) for the Treatment of Pancreatic Cancer.
Acs Med.Chem.Lett., 10, 2019
|
|
9B4E
 
 | | Structure of wild type human PSS1 | | Descriptor: | (2R)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dihexadecanoate, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CALCIUM ION, ... | | Authors: | Long, T, Li, X. | | Deposit date: | 2024-03-20 | | Release date: | 2024-09-04 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Molecular insights into human phosphatidylserine synthase 1 reveal its inhibition promotes LDL uptake.
Cell, 187, 2024
|
|
6RMS
 
 | |
6YQQ
 
 | | ForT-PRPP complex | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, CHLORIDE ION, ForT-PRPP complex, ... | | Authors: | Naismith, J.H, Gao, S. | | Deposit date: | 2020-04-18 | | Release date: | 2020-05-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Uncovering the chemistry of C-C bond formation in C-nucleoside biosynthesis: crystal structure of a C-glycoside synthase/PRPP complex.
Chem.Commun.(Camb.), 56, 2020
|
|
5NX4
 
 | |
8XAM
 
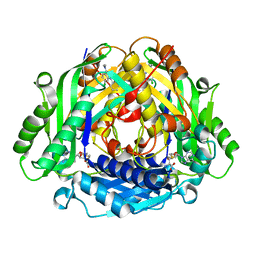 | | Co-crystal structure of compound 7 in complex with MAT2A | | Descriptor: | 2-[3-[7-chloranyl-4-(dimethylamino)-2-oxidanylidene-quinazolin-1-yl]phenoxy]-~{N}-[3-[7-chloranyl-4-(dimethylamino)-2-oxidanylidene-quinazolin-1-yl]phenyl]ethanamide, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Gao, F, Ding, X. | | Deposit date: | 2023-12-04 | | Release date: | 2024-02-28 | | Last modified: | 2025-10-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of novel MAT2A inhibitors by an allosteric site-compatible fragment growing approach.
Bioorg.Med.Chem., 100, 2024
|
|
4Y1U
 
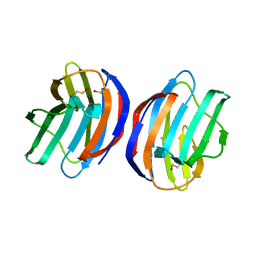 | | Complex of human Galectin-1 and Galbeta1-4GlcNAc | | Descriptor: | Galectin-1, beta-D-galactopyranose-(1-4)-methyl 2-acetamido-2-deoxy-beta-D-glucopyranoside | | Authors: | Lin, H.Y, Hsieh, T.J, Lin, C.H. | | Deposit date: | 2015-02-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.762 Å) | | Cite: | Structural basis of human galectin-1 inhibition with Ki values in the micro- to nanomolar range
To Be Published
|
|
7ATV
 
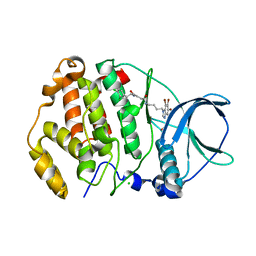 | | Structure of protein kinase ck2 catalytic subunit (csnk2a2 gene product) in complex with the bivalent inhibitor KN2 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Casein kinase II subunit alpha', ... | | Authors: | Lindenblatt, D, Applegate, V, Nickelsen, A, Klussmann, M, Neundorf, I, Goetz, C, Jose, J, Niefind, K. | | Deposit date: | 2020-10-31 | | Release date: | 2021-08-04 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Molecular Plasticity of Crystalline CK2 alpha ' Leads to KN2, a Bivalent Inhibitor of Protein Kinase CK2 with Extraordinary Selectivity.
J.Med.Chem., 65, 2022
|
|
5Y98
 
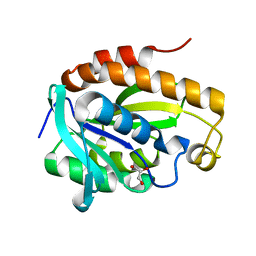 | | Crystal structure of native unbound peptidyl tRNA hydrolase from Acinetobacter baumannii at 1.36 A resolution | | Descriptor: | GLYCEROL, Peptidyl-tRNA hydrolase | | Authors: | Iqbal, N, Singh, N, Kaushik, S, Singh, P.K, Sharma, S, Singh, T.P. | | Deposit date: | 2017-08-23 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Search of multiple hot spots on the surface of peptidyl-tRNA hydrolase: structural, binding and antibacterial studies.
Biochem. J., 475, 2018
|
|
9COP
 
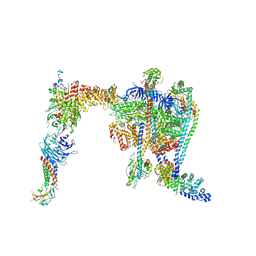 | | Yeast RAVE bound to V-ATPase V1 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Regulator of V-ATPase in vacuolar membrane protein 1, ... | | Authors: | Wang, H, Rubinstein, J.L. | | Deposit date: | 2024-07-17 | | Release date: | 2024-11-20 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure of yeast RAVE bound to a partial V 1 complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SCA
 
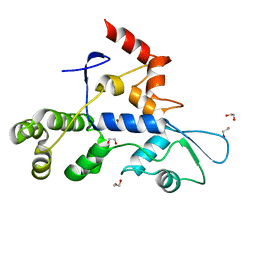 | | Rec3 Domain from S. pyogenes Cas9 | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9/Csn1 | | Authors: | D'Ordine, A.M, Skeens, E, Lisi, G.P, Jogl, G. | | Deposit date: | 2023-04-05 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | High-fidelity, hyper-accurate, and evolved mutants rewire atomic-level communication in CRISPR-Cas9.
Sci Adv, 10, 2024
|
|
5YB2
 
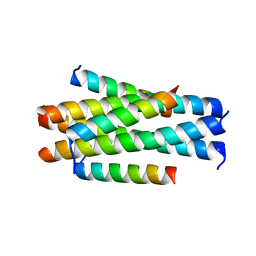 | | Crystal structure of LP-11/N44 | | Descriptor: | Envelope glycoprotein, LP-11 | | Authors: | Zhang, X, Wang, X, He, Y. | | Deposit date: | 2017-09-03 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural Insights into the Mechanisms of Action of Short-Peptide HIV-1 Fusion Inhibitors Targeting the Gp41 Pocket
Front Cell Infect Microbiol, 8, 2018
|
|
5JOE
 
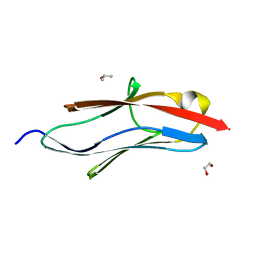 | | Crystal structure of I81 from titin | | Descriptor: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, Titin | | Authors: | Fleming, J, Zhou, T, Bogomolovas, J, Labeit, S, Mayans, O. | | Deposit date: | 2016-05-02 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CARP interacts with titin at a unique helical N2A sequence and at the domain Ig81 to form a structured complex.
Febs Lett., 590, 2016
|
|
6RY5
 
 | | Crystal structure of Dfg5 from Chaetomium thermophilum in complex with alpha-1,6-mannobiose | | Descriptor: | CALCIUM ION, Mannan endo-1,6-alpha-mannosidase, TRIETHYLENE GLYCOL, ... | | Authors: | Essen, L.-O, Vogt, M.S. | | Deposit date: | 2019-06-10 | | Release date: | 2020-08-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural base for the transfer of GPI-anchored glycoproteins into fungal cell walls.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
9ATM
 
 | | SARS-CoV-2 EG.5 RBD bound to the VIR-7229 and the S2H97 Fab fragments | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rietz, T, Park, Y.J, Errico, J, Czudnochowski, N, Nix, J.C, Corti, D, Snell, G, Marco, A.D, Pinto, D, Cameroni, E, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2024-02-27 | | Release date: | 2024-10-16 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A potent pan-sarbecovirus neutralizing antibody resilient to epitope diversification.
Cell, 187, 2024
|
|
