3VQD
 
 | | HIV-1 IN core domain in complex with 5-methyl-3-phenyl-1,2-oxazole-4-carboxylic acid | | Descriptor: | 5-methyl-3-phenyl-1,2-oxazole-4-carboxylic acid, CADMIUM ION, POL polyprotein, ... | | Authors: | Wielens, J, Chalmers, D.K, Parker, M.W, Scanlon, M.J. | | Deposit date: | 2012-03-21 | | Release date: | 2013-01-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Parallel screening of low molecular weight fragment libraries: do differences in methodology affect hit identification?
J Biomol Screen, 18, 2013
|
|
3VQQ
 
 | | HIV-1 integrase core domain in complex with 2,1,3-benzothiadiazol-4-amine | | Descriptor: | 2,1,3-benzothiadiazol-4-amine, CADMIUM ION, POL polyprotein, ... | | Authors: | Wielens, J, Chalmers, D.K, Parker, M.W, Scanlon, M.J. | | Deposit date: | 2012-03-29 | | Release date: | 2013-01-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Parallel screening of low molecular weight fragment libraries: do differences in methodology affect hit identification?
J Biomol Screen, 18, 2013
|
|
8EKB
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with mRNA, deacylated P-site tRNAmet, and thermorubin at 2.70A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Paranjpe, M.N, Polikanov, Y.S. | | Deposit date: | 2022-09-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights into the molecular mechanism of translation inhibition by the ribosome-targeting antibiotic thermorubin.
Nucleic Acids Res., 51, 2023
|
|
4YKP
 
 | |
4RIK
 
 | | Amyloid forming segment, AVVTGVTAV, from the NAC domain of Parkinson's disease protein alpha-synuclein, residues 69-77 | | Descriptor: | Alpha-synuclein | | Authors: | Guenther, E.L, Sawaya, M.R, Ivanova, M, Eisenberg, D.S. | | Deposit date: | 2014-10-06 | | Release date: | 2015-08-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.854 Å) | | Cite: | Structure of the toxic core of alpha-synuclein from invisible crystals.
Nature, 525, 2015
|
|
3VAP
 
 | | Synthesis and SAR Studies of imidazo-[1,2-a]-pyrazine Aurora kinase inhibitors with improved off target kinase selectivity | | Descriptor: | 3-(1-{2-[(3-fluoropyridinium-4-yl)amino]-2-oxoethyl}-1H-pyrazol-4-yl)-6-methyl-8-[(3-{[(1R,3R)-3-methylpiperidinium-1-yl]methyl}-1,2-thiazol-5-yl)amino]imidazo[1,2-a]pyrazin-1-ium, Aurora kinase A | | Authors: | Voss, M.E, Rainka, M.P, Fleming, M, Peterson, L.H, Belanger, D.B, Siddiqui, M.A, Hruza, A, Voigt, J, Basso, A.D, Gray, K. | | Deposit date: | 2011-12-29 | | Release date: | 2012-05-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Synthesis and SAR studies of imidazo-[1,2-a]-pyrazine Aurora kinase inhibitors with improved off-target kinase selectivity.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
5YHS
 
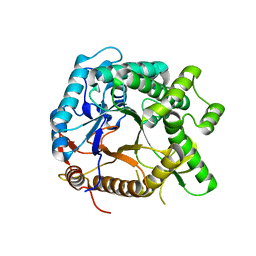 | | Pyruvylated beta-D-galactosidase from Bacillus sp. HMA207, apo form | | Descriptor: | Pyruvylated beta-D-galactosidase | | Authors: | Tanuma, M, Yamada, C, Arakawa, T, Higuchi, Y, Takegawa, K, Fushinobu, S. | | Deposit date: | 2017-09-29 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification and characterization of a novel beta-D-galactosidase that releases pyruvylated galactose.
Sci Rep, 8, 2018
|
|
8EEO
 
 | |
4FGM
 
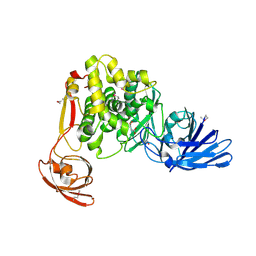 | | Crystal structure of the aminopeptidase N family protein Q5QTY1 from Idiomarina loihiensis. Northeast Structural Genomics Consortium Target IlR60. | | Descriptor: | Aminopeptidase N family protein, MALEIC ACID, ZINC ION | | Authors: | Vorobiev, S, Su, M, Tong, T, Kohan, E, Wang, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-04 | | Release date: | 2012-08-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.394 Å) | | Cite: | Crystal structure of the aminopeptidase N family protein Q5QTY1 from Idiomarina loihiensis.
To be Published
|
|
5C0W
 
 | |
7ZB9
 
 | | Crystal structure of CYP124 in complex with inhibitor carbethoxyhexyl imidazole in the absence of glycerol (NoCryo) | | Descriptor: | CHLORIDE ION, CYP124 in complex with inhibitor carbethoxyhexyl imidazole, MAGNESIUM ION, ... | | Authors: | Bukhdruker, S, Varaksa, T, Marin, E, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2022-03-23 | | Release date: | 2023-01-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural insights into the effects of glycerol on ligand binding to cytochrome P450.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
5Y8V
 
 | | Crystal structure of GAS41 | | Descriptor: | MAGNESIUM ION, YEATS domain-containing protein 4 | | Authors: | Wang, Y, Hao, Q. | | Deposit date: | 2017-08-21 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Identification of the YEATS domain of GAS41 as a pH-dependent reader of histone succinylation
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YIF
 
 | | Pyruvylated beta-D-galactosidase from Bacillus sp. HMA207, E163A mutant pyruvylated beta-D-galactose complex | | Descriptor: | (2R,4aR,6R,7R,8R,8aR)-2-methyl-6,7,8-tris(oxidanyl)-4,4a,6,7,8,8a-hexahydropyrano[3,2-d][1,3]dioxine-2-carboxylic acid, Pyruvylated beta-D-galactosidase | | Authors: | Tanuma, M, Yamada, C, Arakawa, T, Higuchi, Y, Takegawa, K, Fushinobu, S. | | Deposit date: | 2017-10-04 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Identification and characterization of a novel beta-D-galactosidase that releases pyruvylated galactose.
Sci Rep, 8, 2018
|
|
4JWY
 
 | | GluN2D ligand-binding core in complex with propyl-NHP5G | | Descriptor: | (2R)-amino(1-hydroxy-4-propyl-1H-pyrazol-5-yl)ethanoic acid, Glutamate receptor ionotropic, NMDA 2D | | Authors: | Hansen, K.B, Tajima, N, Risgaard, R, Perszyk, R.E, Jorgensen, L, Vance, K.M, Ogden, K.K, Clausen, R.P, Furukawa, H, Traynelis, S.F. | | Deposit date: | 2013-03-27 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural determinants of agonist efficacy at the glutamate binding site of N-methyl-d-aspartate receptors.
Mol.Pharmacol., 84, 2013
|
|
3VQ9
 
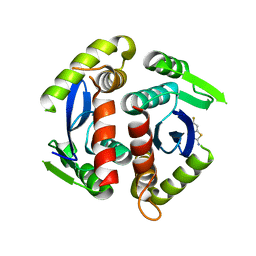 | | HIV-1 IN core domain in complex with 6-fluoro-1,3-benzothiazol-2-amine | | Descriptor: | 6-fluoro-1,3-benzothiazol-2-amine, POL polyprotein | | Authors: | Wielens, J, Chalmers, D.K, Parker, M.W, Scanlon, M.J. | | Deposit date: | 2012-03-20 | | Release date: | 2013-01-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Parallel screening of low molecular weight fragment libraries: do differences in methodology affect hit identification?
J Biomol Screen, 18, 2013
|
|
3VQP
 
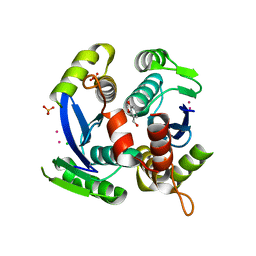 | | HIV-1 IN core domain in complex with 2,3-dihydro-1,4-benzodioxin-5-ylmethanol | | Descriptor: | 2,3-dihydro-1,4-benzodioxin-5-ylmethanol, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Wielens, J, Chalmers, D.K, Parker, M.W, Scanlon, M.J. | | Deposit date: | 2012-03-29 | | Release date: | 2013-01-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Parallel screening of low molecular weight fragment libraries: do differences in methodology affect hit identification?
J Biomol Screen, 18, 2013
|
|
4RHV
 
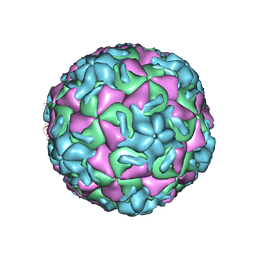 | |
5YKF
 
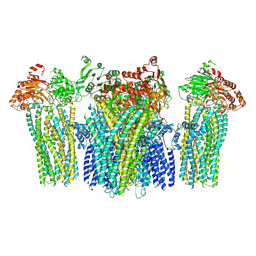 | |
5T7C
 
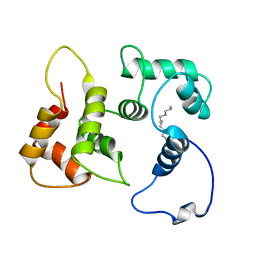 | |
4YKI
 
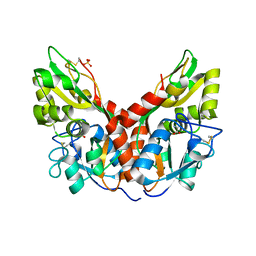 | |
4YKJ
 
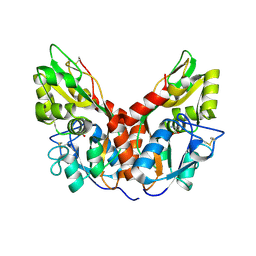 | |
5YTE
 
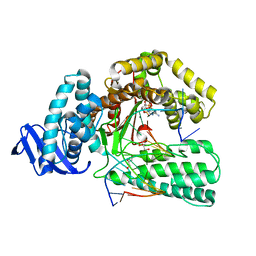 | | Large fragment of DNA Polymerase I from Thermus aquaticus in a closed ternary complex with with natural dT:dATP base pair | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*TP*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), ... | | Authors: | Zeng, H, Mondal, M, Song, R.Y, Zhang, J, Xia, B, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2017-11-17 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Unnatural Cytosine Bases Recognized as Thymines by DNA Polymerases by the Formation of the Watson-Crick Geometry.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
4J76
 
 | | Crystal Structure of a parasite tRNA synthetase, ligand-free | | Descriptor: | GLYCEROL, Tryptophanyl-tRNA synthetase | | Authors: | Koh, C.Y, Kim, J.E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-02-12 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal structures of Plasmodium falciparum cytosolic tryptophanyl-tRNA synthetase and its potential as a target for structure-guided drug design.
Mol.Biochem.Parasitol., 189, 2013
|
|
4RW2
 
 | | Hen egg-white lysozyme structure from a spent-beam experiment at LCLS: refocused beam | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Boutet, S, Foucar, L, Barends, T, Doak, R.B, Koglin, J.E, Messerschmidt, M, Nass, K, Schlichting, I, Shoeman, R, Williams, G.J. | | Deposit date: | 2014-12-01 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization and use of the spent beam for serial operation of LCLS.
J.SYNCHROTRON RADIAT., 22, 2015
|
|
5Y4O
 
 | | Cryo-EM structure of MscS channel, YnaI | | Descriptor: | Low conductance mechanosensitive channel YnaI | | Authors: | Zhang, Y, Yu, J. | | Deposit date: | 2017-08-04 | | Release date: | 2019-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A binding-block ion selective mechanism revealed by a Na/K selective channel.
Protein Cell, 9, 2018
|
|
