2FJZ
 
 | |
1XAL
 
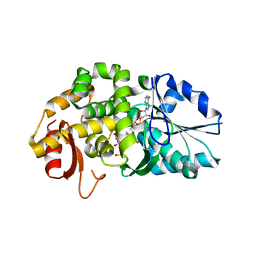 | | CRYSTAL STRUCTURE OF STAPHLYOCOCCUS AUREUS 3-DEHYDROQUINATE SYNTHASE (DHQS) IN COMPLEX WITH ZN2+, NAD+ AND CARBAPHOSPHONATE (SOAK) | | Descriptor: | 3-dehydroquinate synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION, ... | | Authors: | Nichols, C.E, Ren, J, Leslie, K, Dhaliwal, B, Lockyer, M, Charles, I, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2004-08-26 | | Release date: | 2005-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Comparison of ligand induced conformational changes and domain closure mechanisms, between prokaryotic and eukaryotic dehydroquinate synthases.
J.Mol.Biol., 343, 2004
|
|
1VSG
 
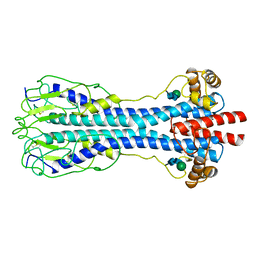 | |
2F55
 
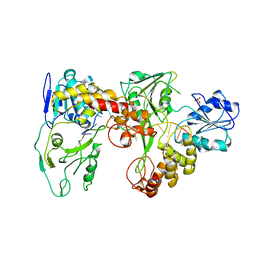 | | Two hepatitis c virus ns3 helicase domains complexed with the same strand of dna | | Descriptor: | 5'-D(P*(DU)P*(DU)P*(DU))-3', 5'-D(P*(DU)P*(DU)P*(DU)P*(DU)P*(DU)P*(DU)P*(DU)P*(DU)P*(DU)P*(DU)P*(DU)P*(DU)P*(DU))-3', SULFATE ION, ... | | Authors: | Lu, J.Z, Jordan, J.B, Sakon, J. | | Deposit date: | 2005-11-25 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural and biological identification of residues on the surface of NS3 helicase required for optimal replication of the hepatitis C virus
J.Biol.Chem., 281, 2006
|
|
1VSO
 
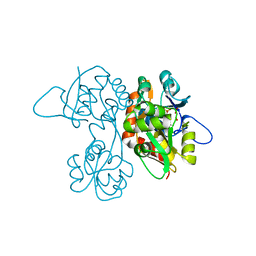 | | Crystal Structure of the Ligand-Binding Core of iGluR5 in Complex With the Antagonist (S)-ATPO at 1.85 A resolution | | Descriptor: | (S)-2-AMINO-3-(5-TERT-BUTYL-3-(PHOSPHONOMETHOXY)-4-ISOXAZOLYL)PROPIONIC ACID, GLYCEROL, Glutamate receptor, ... | | Authors: | Hald, H, Naur, P, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2007-03-29 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Partial agonism and antagonism of the ionotropic glutamate receptor iGLuR5: structures of the ligand-binding core in complex with domoic acid and 2-amino-3-[5-tert-butyl-3-(phosphonomethoxy)-4-isoxazolyl]propionic acid.
J.Biol.Chem., 282, 2007
|
|
1XAG
 
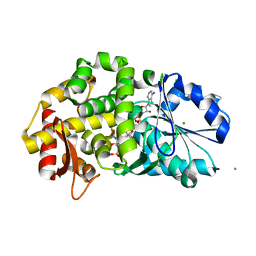 | | CRYSTAL STRUCTURE OF STAPHLYOCOCCUS AUREUS 3-DEHYDROQUINATE SYNTHASE (DHQS) IN COMPLEX WITH ZN2+, NAD+ AND CARBAPHOSPHONATE | | Descriptor: | 3-dehydroquinate synthase, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Nichols, C.E, Ren, J, Leslie, K, Dhaliwal, B, Lockyer, M, Charles, I, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2004-08-25 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Comparison of ligand induced conformational changes and domain closure mechanisms, between prokaryotic and eukaryotic dehydroquinate synthases.
J.Mol.Biol., 343, 2004
|
|
1XDO
 
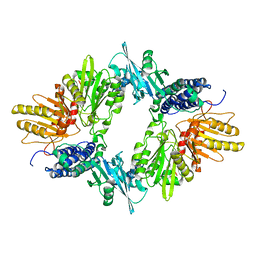 | |
1X7G
 
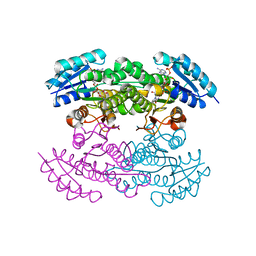 | | Actinorhodin Polyketide Ketoreductase, act KR, with NADP bound | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative ketoacyl reductase | | Authors: | Korman, T.P, Hill, J.A, Vu, T.N. | | Deposit date: | 2004-08-13 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of actinorhodin polyketide ketoreductase: cofactor binding and substrate specificity
Biochemistry, 43, 2004
|
|
1XFD
 
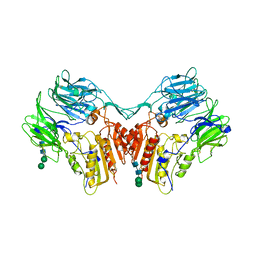 | | Structure of a human A-type Potassium Channel Accelerating factor DPPX, a member of the dipeptidyl aminopeptidase family | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl aminopeptidase-like protein 6, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Strop, P, Bankovich, A.J, Hansen, K.C, Garcia, K.C, Brunger, A.T. | | Deposit date: | 2004-09-14 | | Release date: | 2004-10-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of a human A-type potassium channel interacting protein DPPX, a member of the dipeptidyl aminopeptidase family
J.Mol.Biol., 343, 2004
|
|
1VXO
 
 | |
2EV6
 
 | | Bacillus subtilis manganese transport regulator (MNTR) bound to zinc | | Descriptor: | CITRATE ANION, GLYCEROL, Transcriptional regulator mntR, ... | | Authors: | Kliegman, J.I, Griner, S.L, Helmann, J.D, Brennan, R.G, Glasfeld, A. | | Deposit date: | 2005-10-31 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the Metal-Selective Activation of the Manganese Transport Regulator of Bacillus subtilis.
Biochemistry, 45, 2006
|
|
1VZU
 
 | | Roles of active site tryptophans in substrate binding and catalysis by ALPHA-1,3 GALACTOSYLTRANSFERASE | | Descriptor: | GLYCEROL, MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | Authors: | Zhang, Y, Deshpande, A, Xie, Z, Natesh, R, Acharya, K.R, Brew, K. | | Deposit date: | 2004-05-27 | | Release date: | 2004-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Roles of active site tryptophans in substrate binding and catalysis by alpha-1,3 galactosyltransferase.
Glycobiology, 14, 2004
|
|
1X79
 
 | | Crystal structure of human GGA1 GAT domain complexed with the GAT-binding domain of Rabaptin5 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ADP-ribosylation factor binding protein GGA1, Rab GTPase binding effector protein 1, ... | | Authors: | Zhu, G, Zhang, X.C. | | Deposit date: | 2004-08-13 | | Release date: | 2004-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal structure of human GGA1 GAT domain complexed with the GAT-binding domain of Rabaptin5.
EMBO J., 23, 2004
|
|
1X7R
 
 | | CRYSTAL STRUCTURE OF ESTROGEN RECEPTOR ALPHA COMPLEXED WITH GENISTEIN | | Descriptor: | Estrogen receptor 1 (alpha), GENISTEIN, steroid receptor coactivator-3 | | Authors: | Manas, E.S, Xu, Z.B, Unwalla, R.J, Somers, W.S. | | Deposit date: | 2004-08-16 | | Release date: | 2005-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Understanding the Selectivity of Genistein for Human Estrogen Receptor-Beta Using X-Ray Crystallography and Computational Methods
Structure, 12, 2004
|
|
1W0Z
 
 | | Urokinase type plasminogen activator | | Descriptor: | N-(BUTYLSULFONYL)-D-SERYL-N-{4-[AMINO(IMINO)METHYL]BENZYL}-L-ALANINAMIDE, SULFATE ION, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Jacob, U. | | Deposit date: | 2004-06-15 | | Release date: | 2008-05-20 | | Last modified: | 2019-09-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystals of urokinase type plasminogen activator complexes reveal the binding mode of peptidomimetic inhibitors.
J.Mol.Biol., 328, 2003
|
|
1X8X
 
 | | Tyrosyl t-RNA Synthetase from E.coli Complexed with Tyrosine | | Descriptor: | SULFATE ION, TYROSINE, Tyrosyl-tRNA synthetase | | Authors: | Kobayashi, T, Takimura, T, Sekine, R, Kelly, V.P, Kamata, K, Sakamoto, K, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-08-19 | | Release date: | 2005-01-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Snapshots of the KMSKS Loop Rearrangement for Amino Acid Activation by Bacterial Tyrosyl-tRNA Synthetase
J.MOL.BIOL., 346, 2005
|
|
1VWF
 
 | |
1VDH
 
 | | Structure-based functional identification of a novel heme-binding protein from thermus thermophilus HB8 | | Descriptor: | muconolactone isomerase-like protein | | Authors: | Ebihara, A, Okamoto, A, Kousumi, Y, Yamamoto, H, Masui, R, Ueyama, N, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-22 | | Release date: | 2004-09-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based functional identification of a novel heme-binding protein from Thermus thermophilus HB8.
J.STRUCT.FUNCT.GENOM., 6, 2005
|
|
1VEP
 
 | | Crystal Structure Analysis of Triple (T47M/Y164E/T328N)/maltose of Bacillus cereus Beta-Amylase at pH 6.5 | | Descriptor: | Beta-amylase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Hirata, A, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2004-04-03 | | Release date: | 2005-05-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Engineering of the pH optimum of Bacillus cereus beta-amylase: conversion of the pH optimum from a bacterial type to a higher-plant type
Biochemistry, 43, 2004
|
|
1VTO
 
 | |
1X7H
 
 | | Actinorhodin Polyketide Ketoreductase, with NADPH bound | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative ketoacyl reductase | | Authors: | Korman, T.P, Hill, J.A, Vu, T.N. | | Deposit date: | 2004-08-13 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of actinorhodin polyketide ketoreductase: cofactor binding and substrate specificity
Biochemistry, 43, 2004
|
|
1VZ8
 
 | |
1W01
 
 | |
1W0P
 
 | | Vibrio cholerae sialidase with alpha-2,6-sialyllactose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Moustafa, I, Connaris, H, Taylor, M, Zaitsev, V, Wilson, J.C, Kiefel, M.J, von-Itzstein, M, Taylor, G. | | Deposit date: | 2004-06-09 | | Release date: | 2004-07-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Sialic Acid Recognition by Vibrio Cholerae Neuraminidase.
J.Biol.Chem., 279, 2004
|
|
1W12
 
 | | UROKINASE TYPE PLASMINOGEN ACTIVATOR | | Descriptor: | N-((1S)-4-{[AMINO(IMINO)METHYL]AMINO}-1-FORMYLBUTYL)-2-{(3R)-3-[(BENZYLSULFONYL)AMINO]-2-OXO-5-PHENYL-2,3-DIHYDRO-1H-1,4-BENZODIAZEPIN-1-YL}ACETAMIDE, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Jacob, U. | | Deposit date: | 2004-06-15 | | Release date: | 2008-05-20 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystals of Urokinase Type Plasminogen Activator Complexes Reveal the Binding Mode of Peptidomimetic Inhibitors.
J.Mol.Biol., 328, 2003
|
|
