1P1K
 
 | |
1Q0Q
 
 | | Crystal structure of DXR in complex with the substrate 1-deoxy-D-xylulose-5-phosphate | | Descriptor: | 1-DEOXY-D-XYLULOSE-5-PHOSPHATE, 1-deoxy-D-xylulose 5-phosphate reductoisomerase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Mac Sweeney, A, Lange, R, D'Arcy, A, Douangamath, A, Surivet, J.-P, Oefner, C. | | Deposit date: | 2003-07-17 | | Release date: | 2004-07-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of E.coli 1-deoxy-D-xylulose-5-phosphate reductoisomerase in a ternary complex with the antimalarial compound fosmidomycin and NADPH reveals a tight-binding closed enzyme conformation.
J.Mol.Biol., 345, 2005
|
|
5PGV
 
 | | CRYSTAL STRUCTURE OF 11BETA-HSD1 DOUBLE MUTANT (L262R, F278E) COMPLEXED WITH 1-(3-HYDROXYAZETIDIN-1-YL)-2-[(2S,5R)-2-(4-FLUOROPHENYL)-5-METHOXYADAMANTAN-2-YL]ETHAN-1-ONE | | Descriptor: | 1-(3-HYDROXYAZETIDIN-1-YL)-2-[(2S,5R)-2-(4-FLUOROPHENYL)-5-METHOXYADAMANTAN-2-YL]ETHAN-1-ONE, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sheriff, S. | | Deposit date: | 2017-02-06 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of Clinical Candidate 2-((2S,6S)-2-Phenyl-6-hydroxyadamantan-2-yl)-1-(3'-hydroxyazetidin-1-yl)ethanone [BMS-816336], an Orally Active Novel Selective 11 beta-Hydroxysteroid Dehydrogenase Type 1 Inhibitor.
J. Med. Chem., 60, 2017
|
|
1P1J
 
 | | Crystal structure of the 1L-myo-inositol 1-phosphate synthase complexed with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, Inositol-3-phosphate synthase, ... | | Authors: | Jin, X, Geiger, J.H. | | Deposit date: | 2003-04-12 | | Release date: | 2003-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of NAD(+)- and NADH-bound 1-l-myo-inositol 1-phosphate synthase.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1I75
 
 | | CRYSTAL STRUCTURE OF CYCLODEXTRIN GLUCANOTRANSFERASE FROM ALKALOPHILIC BACILLUS SP.#1011 COMPLEXED WITH 1-DEOXYNOJIRIMYCIN | | Descriptor: | 1-DEOXYNOJIRIMYCIN, CALCIUM ION, CYCLODEXTRIN GLUCANOTRANSFERASE | | Authors: | Kanai, R, Haga, K, Yamane, K, Harata, K. | | Deposit date: | 2001-03-08 | | Release date: | 2001-04-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of cyclodextrin glucanotransferase from alkalophilic Bacillus sp. 1011 complexed with 1-deoxynojirimycin at 2.0 A resolution.
J.Biochem.(Tokyo), 129, 2001
|
|
7DE5
 
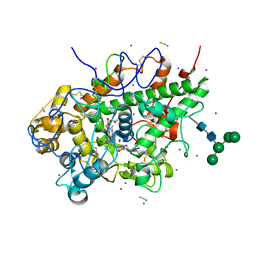 | | Crystal structure of yak lactoperoxidase at 1.55 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, P.K, Viswanathan, V, Sharma, P, Rani, C, Ahmad, N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2020-11-02 | | Release date: | 2020-11-25 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of Yak Lactoperoxidase at 1.55 angstrom Resolution.
Protein J., 40, 2021
|
|
1G6L
 
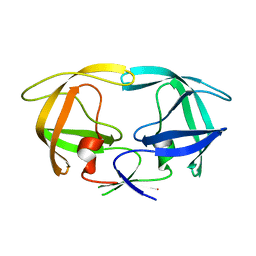 | |
1JKI
 
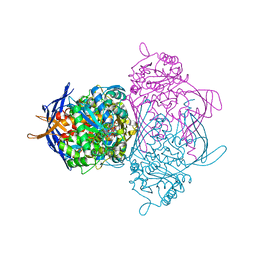 | | myo-Inositol-1-phosphate Synthase Complexed with an Inhibitor, 2-deoxy-glucitol-6-phosphate | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-DEOXY-GLUCITOL-6-PHOSPHATE, AMMONIUM ION, ... | | Authors: | Stein, A.J, Geiger, J.H. | | Deposit date: | 2001-07-12 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure and mechanism of 1-L-myo-inositol- 1-phosphate synthase
J.Biol.Chem., 277, 2002
|
|
2AZQ
 
 | | Crystal Structure of Catechol 1,2-Dioxygenase from Pseudomonas arvilla C-1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, FE (III) ION, catechol 1,2-dioxygenase | | Authors: | Earhart, C.A, Vetting, M.W, Gosu, R, Michaud-Soret, I, Que, L, Ohlendorf, D.H. | | Deposit date: | 2005-09-12 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of catechol 1,2-dioxygenase from Pseudomonas arvilla
Biochem.Biophys.Res.Commun., 338, 2005
|
|
2DOO
 
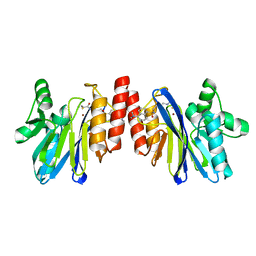 | | The structure of IMP-1 complexed with the detecting reagent (DansylC4SH) by a fluorescent probe | | Descriptor: | BETA-LACTAMASE IMP-1, N-[4-({[5-(DIMETHYLAMINO)-1-NAPHTHYL]SULFONYL}AMINO)BUTYL]-3-SULFANYLPROPANAMIDE, ZINC ION | | Authors: | Kurosaki, H, Yamaguchi, Y, Yasuzawa, H, Jin, W, Yamagata, Y, Arakawa, Y. | | Deposit date: | 2006-05-01 | | Release date: | 2006-11-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Probing, inhibition, and crystallographic characterization of metallo-beta-lactamase (IMP-1) with fluorescent agents containing dansyl and thiol groups
Chemmedchem, 1, 2006
|
|
3RAS
 
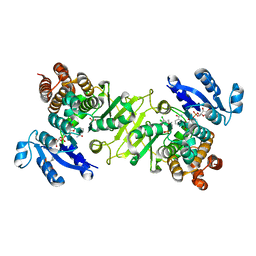 | | Crystal structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase (DXR) complexed with a lipophilic phosphonate inhibitor | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-(N-HYDROXYACETAMIDO)-1-(3,4-DICHLOROPHENYL)PROPYLPHOSPHONIC ACID, MANGANESE (II) ION, ... | | Authors: | Diao, J, Deng, L, Prasad, B.V.V, Song, Y. | | Deposit date: | 2011-03-28 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Inhibition of 1-deoxy-D-xylulose-5-phosphate reductoisomerase by lipophilic phosphonates: SAR, QSAR, and crystallographic studies.
J.Med.Chem., 54, 2011
|
|
2Q6N
 
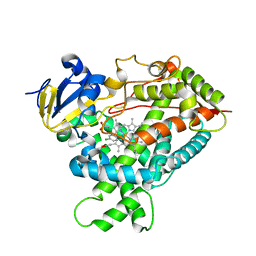 | | Structure of Cytochrome P450 2B4 with Bound 1-(4-cholorophenyl)imidazole | | Descriptor: | 1-(4-CHLOROPHENYL)-1H-IMIDAZOLE, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Zhao, Y, Sun, L, Muralidhara, B.K, Kumar, S, White, M.A, Stout, C.D, Halpert, J.R. | | Deposit date: | 2007-06-05 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and thermodynamic consequences of 1-(4-chlorophenyl)imidazole binding to cytochrome P450 2B4.
Biochemistry, 46, 2007
|
|
2OO5
 
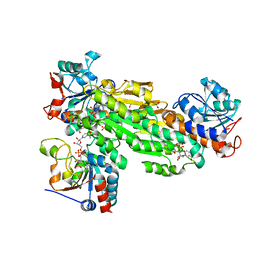 | | Structure of transhydrogenase (dI.H2NADH)2(dIII.NADP+)1 asymmetric complex | | Descriptor: | 1,4,5,6-TETRAHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, NAD(P) transhydrogenase subunit alpha part 1, NAD(P) transhydrogenase subunit beta, ... | | Authors: | Bhakta, T, Jackson, J.B. | | Deposit date: | 2007-01-25 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of the dI(2)dIII(1) Complex of Proton-Translocating Transhydrogenase with Bound, Inactive Analogues of NADH and NADPH Reveal Active Site Geometries
Biochemistry, 46, 2007
|
|
4I42
 
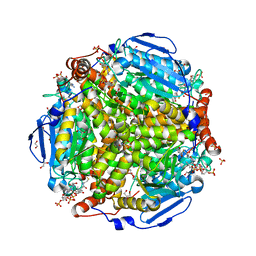 | | E.coli. 1,4-dihydroxy-2-naphthoyl coenzyme A synthase (ecMenB) in complex with 1-hydroxy-2-naphthoyl-CoA | | Descriptor: | 1,2-ETHANEDIOL, 1,4-Dihydroxy-2-naphthoyl-CoA synthase, 1-hydroxy-2-naphthoyl-CoA, ... | | Authors: | Sun, Y, Song, H, Li, J, Li, Y, Jiang, M, Zhou, J, Guo, Z. | | Deposit date: | 2012-11-27 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Structural basis of the induced-fit mechanism of 1,4-dihydroxy-2-naphthoyl coenzyme A synthase from the crotonase fold superfamily
Plos One, 8, 2013
|
|
4K26
 
 | | 4,4-Dioxo-5,6-dihydro-[1,4,3]oxathiazines, a novel class of 11 -HSD1 inhibitors for the treatment of diabetes | | Descriptor: | (4aS,8aR)-N-cyclohexyl-4a,5,6,7,8,8a-hexahydro-4,1,2-benzoxathiazin-3-amine 1,1-dioxide, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Loenze, P, Schimanski-Breves, S, Vonderheyden, C, Engel, C.K. | | Deposit date: | 2013-04-08 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | 1,1-Dioxo-5,6-dihydro-[4,1,2]oxathiazines, a novel class of 11-HSD1 inhibitors for the treatment of diabetes.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4K1L
 
 | | 4,4-Dioxo-5,6-dihydro-[1,4,3]oxathiazines, a novel class of 11 beta-HSD1 inhibitors for the treatment of diabetes | | Descriptor: | (4aS,8aR)-N-cyclohexyl-4a,5,6,7,8,8a-hexahydro-4,1,2-benzoxathiazin-3-amine 1,1-dioxide, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Loenze, P, Schimanski-Breves, S, Von der Heyden, C, Engel, C.K. | | Deposit date: | 2013-04-05 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | 1,1-Dioxo-5,6-dihydro-[4,1,2]oxathiazines, a novel class of 11-HSD1 inhibitors for the treatment of diabetes.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
2OOR
 
 | | Structure of transhydrogenase (dI.NAD+)2(dIII.H2NADPH)1 asymmetric complex | | Descriptor: | 1,4,5,6-TETRAHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE PHOSPHATE, GLYCEROL, NAD(P) transhydrogenase subunit alpha part 1, ... | | Authors: | Bhakta, T, Jackson, J.B. | | Deposit date: | 2007-01-26 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structures of the dI(2)dIII(1) Complex of Proton-Translocating Transhydrogenase with Bound, Inactive Analogues of NADH and NADPH Reveal Active Site Geometries
Biochemistry, 46, 2007
|
|
2EAB
 
 | | Crystal structure of 1,2-a-L-fucosidase from Bifidobacterium bifidum (apo form) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-fucosidase, ... | | Authors: | Nagae, M, Tsuchiya, A, Katayama, T, Yamamoto, K, Wakatsuki, S, Kato, R. | | Deposit date: | 2007-01-31 | | Release date: | 2007-04-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structural basis on the catalytic reaction mechanism of novel 1,2-alpha-L-fucosidase (AFCA) from Bifidobacterium bifidum
J.Biol.Chem., 282, 2007
|
|
1E2K
 
 | | Kinetics and crystal structure of the wild-type and the engineered Y101F mutant of Herpes simplex virus type 1 thymidine kinase interacting with (North)-methanocarba-thymidine | | Descriptor: | 1-[4-HYDROXY-5-(HYDROXYMETHYL)BICYCLO[3.1.0]HEX-2-YL]-5-METHYLPYRIMIDINE-2,4(1H,3H)-DIONE, SULFATE ION, THYMIDINE KINASE | | Authors: | Vogt, J, Scapozza, L, Schulz, G.E. | | Deposit date: | 2000-05-23 | | Release date: | 2000-08-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Kinetics and Crystal Structure of the Wild-Type and the Engineered Y101F Mutant of Herpes Simplex Virus Type 1 Thymidine Kinase Interacting with (North)-Methanocarba-Thymidine
Biochemistry, 39, 2000
|
|
1E2L
 
 | | Kinetics and crystal structure of the wild-type and the engineered Y101F mutant of Herpes simplex virus type 1 thymidine kinase interacting with (North)-methanocarba-thymidine | | Descriptor: | 1-[4-HYDROXY-5-(HYDROXYMETHYL)BICYCLO[3.1.0]HEX-2-YL]-5-METHYLPYRIMIDINE-2,4(1H,3H)-DIONE, SULFATE ION, THYMIDINE KINASE | | Authors: | Vogt, J, Scapozza, L, Schulz, G.E. | | Deposit date: | 2000-05-23 | | Release date: | 2000-08-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Kinetics and Crystal Structure of the Wild-Type and the Engineered Y101F Mutant of Herpes Simplex Virus Type 1 Thymidine Kinase Interacting with (North)-Methanocarba-Thymidine
Biochemistry, 39, 2000
|
|
8XUC
 
 | | Crystal structure of AtHPPD + ZJ-1 complex | | Descriptor: | (1~{S},5~{R})-3-(3-chloranyl-1-ethyl-pyrrolo[2,3-b]pyridin-4-yl)carbonyl-4-oxidanyl-bicyclo[3.2.1]oct-3-en-2-one, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2024-01-12 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of AtHPPD + ZJ-1 complex
To Be Published
|
|
2SRT
 
 | |
1FLI
 
 | | DNA-BINDING DOMAIN OF FLI-1 | | Descriptor: | FLI-1 | | Authors: | Liang, H, Mao, X, Olejniczak, E.T, Nettesheim, D.G, Yu, L, Meadows, R.P, Thompson, C.B, Fesik, S.W. | | Deposit date: | 1994-09-15 | | Release date: | 1995-09-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ets domain of Fli-1 when bound to DNA.
Nat.Struct.Biol., 1, 1994
|
|
6FY4
 
 | |
8IRG
 
 | | XFEL structure of cyanobacterial photosystem II following two flashes (2F) with a 30-microsecond delay | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, H, Suga, M, Shen, J.R. | | Deposit date: | 2023-03-17 | | Release date: | 2024-01-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Oxygen-evolving photosystem II structures during S 1 -S 2 -S 3 transitions.
Nature, 626, 2024
|
|
