1BG2
 
 | | HUMAN UBIQUITOUS KINESIN MOTOR DOMAIN | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, KINESIN, ... | | Authors: | Kull, F.J, Sablin, E.P, Lau, R, Fletterick, R.J, Vale, R.D. | | Deposit date: | 1998-06-04 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the kinesin motor domain reveals a structural similarity to myosin.
Nature, 380, 1996
|
|
1BC1
 
 | | RECOMBINANT RAT ANNEXIN V, QUADRUPLE MUTANT (T72K, S144K, S228K, S303K) | | Descriptor: | ANNEXIN V, CALCIUM ION, SULFATE ION | | Authors: | Mo, Y.D, Swairjo, M.A, Li, C.W, Head, J.F, Seaton, B.A. | | Deposit date: | 1998-05-04 | | Release date: | 1998-11-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mutational and crystallographic analyses of interfacial residues in annexin V suggest direct interactions with phospholipid membrane components.
Biochemistry, 37, 1998
|
|
1BCZ
 
 | | RECOMBINANT RAT ANNEXIN V, T72S MUTANT | | Descriptor: | ANNEXIN V, CALCIUM ION, SULFATE ION | | Authors: | Mo, Y.D, Swairjo, M.A, Li, C.W, Head, J.F, Seaton, B.A. | | Deposit date: | 1998-05-04 | | Release date: | 1998-11-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutational and crystallographic analyses of interfacial residues in annexin V suggest direct interactions with phospholipid membrane components.
Biochemistry, 37, 1998
|
|
1BCW
 
 | | RECOMBINANT RAT ANNEXIN V, T72A MUTANT | | Descriptor: | ANNEXIN V, CALCIUM ION, SULFATE ION | | Authors: | Mo, Y.D, Swairjo, M.A, Li, C.W, Head, J.F, Seaton, B.A. | | Deposit date: | 1998-05-04 | | Release date: | 1998-11-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mutational and crystallographic analyses of interfacial residues in annexin V suggest direct interactions with phospholipid membrane components.
Biochemistry, 37, 1998
|
|
1ZXN
 
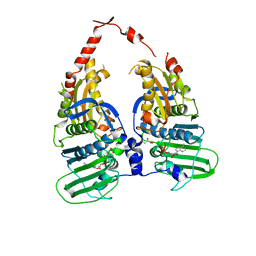 | | Human DNA topoisomerase IIa ATPase/ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA topoisomerase II, alpha isozyme, ... | | Authors: | Wei, H, Ruthenburg, A.J, Bechis, S.K, Verdine, G.L. | | Deposit date: | 2005-06-08 | | Release date: | 2005-08-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Nucleotide-dependent Domain Movement in the ATPase Domain of a Human Type IIA DNA Topoisomerase.
J.Biol.Chem., 280, 2005
|
|
1DXO
 
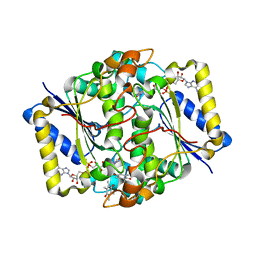 | | Crystal structure of human NAD[P]H-QUINONE oxidoreductase CO with 2,3,5,6,tetramethyl-P-benzoquinone (duroquinone) at 2.5 Angstrom resolution | | Descriptor: | DUROQUINONE, FLAVIN-ADENINE DINUCLEOTIDE, QUINONE REDUCTASE | | Authors: | Faig, M, Bianchet, M.A, Chen, S, Winski, S, Ross, D, Amzel, L.M. | | Deposit date: | 2000-01-12 | | Release date: | 2000-04-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Recombinant Mouse and Human Nad(P)H:Quinone Oxidoreductases:Species Comparison and Structural Changes with Substrate Binding and Release
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1DXQ
 
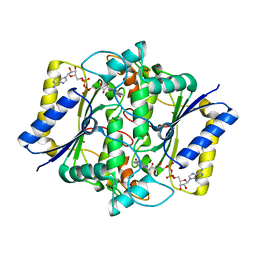 | | CRYSTAL STRUCTURE OF MOUSE NAD[P]H-QUINONE OXIDOREDUCTASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, QUINONE REDUCTASE | | Authors: | Faig, M, Bianchet, M.A, Chen, S, Winski, S, Ross, D, Amzel, L.M. | | Deposit date: | 2000-01-14 | | Release date: | 2000-04-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of Recombinant Mouse and Human Nad(P)H:Quinone Oxidoreductases:Species Comparison and Structural Changes with Substrate Binding and Release
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
2ADB
 
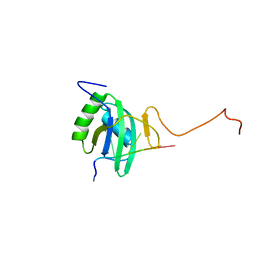 | | Solution structure of Polypyrimidine Tract Binding protein RBD2 complexed with CUCUCU RNA | | Descriptor: | 5'-R(*CP*UP*CP*UP*CP*U)-3', Polypyrimidine tract-binding protein 1 | | Authors: | Oberstrass, F.C, Auweter, S.D, Erat, M, Hargous, Y, Henning, A, Wenter, P, Reymond, L, Pitsch, S, Black, D.L, Allain, F.H.T. | | Deposit date: | 2005-07-20 | | Release date: | 2005-10-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of PTB bound to RNA: specific binding and implications for splicing regulation
Science, 309, 2005
|
|
1DZ1
 
 | | Mouse HP1 (M31) C terminal (shadow chromo) domain | | Descriptor: | MODIFIER 1 PROTEIN | | Authors: | Brasher, S.V, Smith, B.O, Fogh, R.H, Nietlispach, D, Thiru, A, Nielsen, P.R, Broadhurst, R.W, Ball, L.J, Murzina, N, Laue, E.D. | | Deposit date: | 2000-02-11 | | Release date: | 2000-04-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Structure of Mouse Hp1 Suggests a Unique Mode of Single Peptide Recognition by the Shadow Chromo Domain Dimer
Embo J., 19, 2000
|
|
2AKZ
 
 | | Fluoride Inhibition of Enolase: Crystal Structure of the Inhibitory Complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FLUORIDE ION, Gamma enolase, ... | | Authors: | Qin, J, Chai, G, Brewer, J.M, Lovelace, L.L. | | Deposit date: | 2005-08-04 | | Release date: | 2006-03-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Fluoride inhibition of enolase: crystal structure and thermodynamics
Biochemistry, 45, 2006
|
|
2AZM
 
 | | Crystal structure of the MDC1 brct repeat in complex with the histone tail of gamma-H2AX | | Descriptor: | GAMMA-H2AX HISTONE, Mediator of DNA damage checkpoint protein 1 | | Authors: | Clapperton, J.A, Stucki, M, Mohammad, D, Yaffe, M.B, Jackson, S.P, Smerdon, S.J. | | Deposit date: | 2005-09-12 | | Release date: | 2006-01-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | MDC1 Directly Binds Phosphorylated Histone H2AX to Regulate Cellular Responses to DNA Double-Strand Breaks
Cell(Cambridge,Mass.), 123, 2005
|
|
1CJY
 
 | | HUMAN CYTOSOLIC PHOSPHOLIPASE A2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, PROTEIN (CYTOSOLIC PHOSPHOLIPASE A2) | | Authors: | Dessen, A, Tang, J, Schmidt, H, Stahl, M, Clark, J.D, Seehra, J, Somers, W.S. | | Deposit date: | 1999-04-20 | | Release date: | 2000-04-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human cytosolic phospholipase A2 reveals a novel topology and catalytic mechanism.
Cell(Cambridge,Mass.), 97, 1999
|
|
2ALF
 
 | | crystal structure of human CypA mutant K131A | | Descriptor: | MAGNESIUM ION, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Hu, H, Huang, C.-Q, Liu, H.-L, Han, Y, Chen, M.-E, Yu, L, Bi, R.-C. | | Deposit date: | 2005-08-05 | | Release date: | 2005-08-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nuclease activity of Cyclophilin A and its structural basis
TO BE PUBLISHED
|
|
2AKM
 
 | | Fluoride Inhibition of Enolase: Crystal Structure of the Inhibitory Complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Gamma enolase, MAGNESIUM ION, ... | | Authors: | Qin, J, Chai, G, Brewer, J.M, Lovelace, L.L. | | Deposit date: | 2005-08-03 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Fluoride inhibition of enolase: crystal structure and thermodynamics
Biochemistry, 45, 2006
|
|
2B8A
 
 | | High Resolution Structure of the HDGF PWWP Domain | | Descriptor: | Hepatoma-derived growth factor | | Authors: | Lukasik, S.M, Cierpicki, T, Borloz, M, Grembecka, J, Everett, A, Bushweller, J.H. | | Deposit date: | 2005-10-06 | | Release date: | 2005-12-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High resolution structure of the HDGF PWWP domain: a potential DNA binding domain.
Protein Sci., 15, 2006
|
|
1EJF
 
 | | CRYSTAL STRUCTURE OF THE HUMAN CO-CHAPERONE P23 | | Descriptor: | Prostaglandin E synthase 3, SULFATE ION | | Authors: | Weaver, A.J, Sullivan, W.P, Felts, S.J, Owen, B.A.L, Toft, D.O. | | Deposit date: | 2000-03-02 | | Release date: | 2000-06-19 | | Last modified: | 2020-10-28 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structure and activity of human p23, a heat shock protein 90 co-chaperone.
J.Biol.Chem., 275, 2000
|
|
1EWE
 
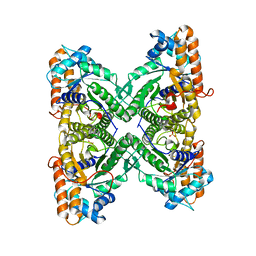 | | Fructose 1,6-Bisphosphate Aldolase from Rabbit Muscle | | Descriptor: | FRUCTOSE 1,6-BISPHOSPHATE ALDOLASE, SULFATE ION | | Authors: | Maurady, A, Sygusch, J. | | Deposit date: | 2000-04-25 | | Release date: | 2001-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A conserved glutamate residue exhibits multifunctional catalytic roles in
D-fructose-1,6-bisphosphate aldolases.
J.Biol.Chem., 277, 2002
|
|
5I80
 
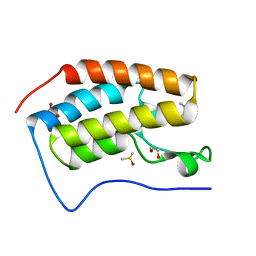 | | BRD4 in complex with Cpd2 (N,N-dimethyl-3-(6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl)benzamide) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, DIMETHYL SULFOXIDE, ... | | Authors: | Murray, J.M. | | Deposit date: | 2016-02-18 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.4501 Å) | | Cite: | Diving into the Water: Inducible Binding Conformations for BRD4, TAF1(2), BRD9, and CECR2 Bromodomains.
J.Med.Chem., 59, 2016
|
|
5IDZ
 
 | |
3I7O
 
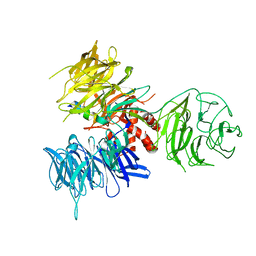 | | Crystal Structure of DDB1 in Complex with the H-Box Motif of IQWD1 | | Descriptor: | DNA damage-binding protein 1, IQ motif and WD repeat-containing protein 1 | | Authors: | Li, T, Robert, E.I, Breugel, P.C.V, Strubin, M, Zheng, N. | | Deposit date: | 2009-07-08 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery.
Nat.Struct.Mol.Biol., 17, 2010
|
|
5IFS
 
 | | Quantitative interaction mapping reveals an extended ubiquitin regulatory domain in ASPL that disrupts functional p97 hexamers and induces cell death | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Roske, Y, Arumughan, A, Heinemann, U, Wanker, E. | | Deposit date: | 2016-02-26 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Quantitative interaction mapping reveals an extended UBX domain in ASPL that disrupts functional p97 hexamers.
Nat Commun, 7, 2016
|
|
5IIT
 
 | |
3IHI
 
 | | Crystal structure of mouse thymidylate synthase | | Descriptor: | SULFATE ION, Thymidylate synthase | | Authors: | Dowiercial, A, Jarmula, A, Rypniewski, W.R, Sokolowska, M, Fraczyk, T, Ciesla, J, Rode, W. | | Deposit date: | 2009-07-30 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mouse thymidylate synthase does not show the inactive conformation, observed for the human enzyme
Struct Chem, 2016
|
|
5ICN
 
 | | HDAC1:MTA1 in complex with inositol-6-phosphate and a novel peptide inhibitor based on histone H4 | | Descriptor: | GLY-ALA-6A0-ARG-HIS, Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Millard, C.J, Robertson, N.S, Watson, P.J, Jameson, A.G, Schwabe, J.W.R. | | Deposit date: | 2016-02-23 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Insights into the activation mechanism of class I HDAC complexes by inositol phosphates.
Nat Commun, 7, 2016
|
|
7ZIR
 
 | | Cryo-EM structure of hnRNPDL amyloid fibrils | | Descriptor: | Heterogeneous nuclear ribonucleoprotein D-like | | Authors: | Garcia-Pardo, J, Chaves-Sanjuan, A, Bartolome-Nafria, A, Gil-Garcia, M, Visentin, C, Bolognesi, M, Ricagno, S, Ventura, S. | | Deposit date: | 2022-04-08 | | Release date: | 2022-12-28 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structure of hnRNPDL-2 fibrils, a functional amyloid associated with limb-girdle muscular dystrophy D3.
Nat Commun, 14, 2023
|
|
