8HE3
 
 | |
3TV3
 
 | |
9LRR
 
 | | Cryo-EM structure of Na+-translocating NADH-ubiquinone oxidoreductase NqrB-G141A mutant from Vibrio cholerae with bound korormicin A | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Ishikawa-Fukuda, M, Kishikawa, J, Kato, T, Murai, M. | | Deposit date: | 2025-02-01 | | Release date: | 2025-04-23 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structural Elucidation of the Mechanism for Inhibitor Resistance in the Na + -Translocating NADH-Ubiquinone Oxidoreductase from Vibrio cholerae.
Biochemistry, 64, 2025
|
|
8I1C
 
 | |
9LVR
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with compound 1 | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, 6-(1,3-dihydroisoindol-2-yl)-3-(5-methylpyridin-3-yl)-1-[[3,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Unoh, Y, Hirai, K, Uehara, S, Kawashima, S, Nobori, H, Sato, J, Shibayama, H, Hori, A, Nakahara, K, Kurahashi, K, Takamatsu, M, Yamamoto, S, Zhang, O, Tanimura, M, Dodo, R, Maruyama, Y, Sawa, H, Watari, R, Miyano, T, Kato, T, Sato, T, Tachibana, Y. | | Deposit date: | 2025-02-12 | | Release date: | 2025-05-14 | | Last modified: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of the Clinical Candidate S-892216 : A Second-Generation of SARS-CoV-2 3CL Protease Inhibitor for Treating COVID-19.
J.Med.Chem., 2025
|
|
8I1G
 
 | |
4Y69
 
 | | Yeast 20S proteasome in complex with Ac-PAD-ep | | Descriptor: | Ac-PAD-ep, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-02-12 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors.
J.Am.Chem.Soc., 137, 2015
|
|
4EV4
 
 | | Crystal structure of serratia fonticola carbapenemase SFC-1 E166A mutant with the acylenzyme intermediate of meropenem | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, 1,2-ETHANEDIOL, Carbapenem-hydrolizing beta-lactamase SFC-1 | | Authors: | Fonseca, F, Spencer, J. | | Deposit date: | 2012-04-25 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The basis for carbapenem hydrolysis by class A beta-lactamases: a combined investigation using crystallography and simulations.
J.Am.Chem.Soc., 134, 2012
|
|
8HBF
 
 | |
1TK4
 
 | | Crystal structure of russells viper phospholipase A2 in complex with a specifically designed tetrapeptide Ala-Ile-Arg-Ser at 1.1 A resolution | | Descriptor: | Phospholipase A2 VRV-PL-VIIIa, SULFATE ION, Tetrapeptide Ala-Ile-Arg-Ser | | Authors: | Singh, N, Bilgrami, S, Somvanshi, R.K, Sharma, S, Dey, S, Perbandt, M, Betzel, C, Kaur, P, Singh, T.P. | | Deposit date: | 2004-06-08 | | Release date: | 2004-06-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structure of russells viper phospholipase A2 with a specifically designed tetrapeptide Ala-Ile-Arg-Ser at 1.1 A resolution
TO BE PUBLISHED
|
|
7VUO
 
 | |
4EA2
 
 | | Crystal structure of dehydrosqualene synthase (Crtm) aureus complexed with SQ-109 | | Descriptor: | Dehydrosqualene synthase, MAGNESIUM ION, N-[(2Z)-3,7-dimethylocta-2,6-dien-1-yl]-N'-[(1R,3S,5R,7R)-tricyclo[3.3.1.1~3,7~]dec-2-yl]ethane-1,2-diamine, ... | | Authors: | Lin, F.-Y, Li, K, Liu, Y.-L, Oldfield, E. | | Deposit date: | 2012-03-21 | | Release date: | 2012-04-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Head-to-Head Prenyl Tranferases: Anti-Infective Drug Targets.
J.Med.Chem., 55, 2012
|
|
1TLC
 
 | | THYMIDYLATE SYNTHASE COMPLEXED WITH DGMP AND FOLATE ANALOG 1843U89 | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, S)-2-(5(((1,2-DIHYDRO-3-METHYL-1-OXOBENZO(F)QUINAZOLIN-9-YL)METHYL)AMINO)1-OXO-2-ISOINDOLINYL)GLUTARIC ACID, THYMIDYLATE SYNTHASE | | Authors: | Weichsel, A, Montfort, W.R, Ciesla, J, Maley, F. | | Deposit date: | 1995-03-07 | | Release date: | 1995-06-03 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Promotion of purine nucleotide binding to thymidylate synthase by a potent folate analogue inhibitor, 1843U89.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1TBC
 
 | |
6NRH
 
 | | Crystal Structure of human PARP-1 ART domain bound inhibitor UTT63 | | Descriptor: | 3-hydroxy-2-({4-[4-(pyrimidin-2-yl)piperazine-1-carbonyl]phenyl}methyl)-1-benzofuran-7-carboxamide, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 1, ... | | Authors: | Langelier, M.F, Pascal, J.M. | | Deposit date: | 2019-01-23 | | Release date: | 2019-08-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design and Synthesis of Poly(ADP-ribose) Polymerase Inhibitors: Impact of Adenosine Pocket-Binding Motif Appendage to the 3-Oxo-2,3-dihydrobenzofuran-7-carboxamide on Potency and Selectivity.
J.Med.Chem., 62, 2019
|
|
8H9S
 
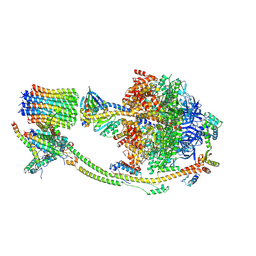 | | Human ATP synthase state 1 (combined) | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Lai, Y, Zhang, Y, Liu, F, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2022-10-25 | | Release date: | 2023-05-31 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structure of the human ATP synthase.
Mol.Cell, 83, 2023
|
|
1TCW
 
 | | SIV PROTEASE COMPLEXED WITH INHIBITOR SB203386 | | Descriptor: | (2R,4S,5S,1'S)-2-PHENYLMETHYL-4-HYDROXY-5-(TERT-BUTOXYCARBONYL)AMINO-6-PHENYL HEXANOYL-N-(1'-IMIDAZO-2-YL)-2'-METHYLPROPANAMIDE, SIV PROTEASE | | Authors: | Hoog, S.S, Abdel-Meguid, S.S. | | Deposit date: | 1996-06-05 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human immunodeficiency virus protease ligand specificity conferred by residues outside of the active site cavity.
Biochemistry, 35, 1996
|
|
1THE
 
 | |
1SUP
 
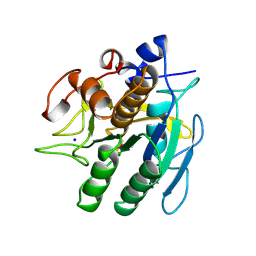 | | SUBTILISIN BPN' AT 1.6 ANGSTROMS RESOLUTION: ANALYSIS OF DISCRETE DISORDER AND COMPARISON OF CRYSTAL FORMS | | Descriptor: | CALCIUM ION, SODIUM ION, SUBTILISIN BPN', ... | | Authors: | Gallagher, D.T, Oliver, J.D, Betzel, C, Gilliland, G.L. | | Deposit date: | 1995-08-14 | | Release date: | 1995-11-14 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Subtilisin BPN' at 1.6 A resolution: analysis for discrete disorder and comparison of crystal forms.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
4Y7X
 
 | | Yeast 20S proteasome in complex with Ac-PAA-ep | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ac-PPA-ep, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-02-16 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors.
J.Am.Chem.Soc., 137, 2015
|
|
4Y8Z
 
 | |
6NRJ
 
 | | Crystal Structure of human PARP-1 ART domain bound to inhibitor UTT93 | | Descriptor: | (2Z)-2-[(4-{[2-(1H-benzimidazol-2-yl)ethyl]carbamoyl}phenyl)methylidene]-3-oxo-2,3-dihydro-1-benzofuran-7-carboxamide, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 1, ... | | Authors: | Langelier, M.F, Pascal, J.M. | | Deposit date: | 2019-01-23 | | Release date: | 2019-08-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Design and Synthesis of Poly(ADP-ribose) Polymerase Inhibitors: Impact of Adenosine Pocket-Binding Motif Appendage to the 3-Oxo-2,3-dihydrobenzofuran-7-carboxamide on Potency and Selectivity.
J.Med.Chem., 62, 2019
|
|
7VWM
 
 | | Crystal Structure of the Y53F/N55A/I116V mutant of LEH | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Qu, G, Li, X, Sun, Z.T, Han, X, Liu, W.D. | | Deposit date: | 2021-11-11 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Rational enzyme design for enabling biocatalytic Baldwin cyclization and asymmetric synthesis of chiral heterocycles.
Nat Commun, 13, 2022
|
|
9CCJ
 
 | |
4Y7B
 
 | | Factor Xa complex with GTC000441 | | Descriptor: | 6-chloro-N-{(3S)-1-[(2S)-1-{(1R,5S)-7-[2-(methylamino)ethyl]-3,7-diazabicyclo[3.3.1]non-3-yl}-1-oxopropan-2-yl]-2-oxopy rrolidin-3-yl}naphthalene-2-sulfonamide, CALCIUM ION, Coagulation factor X | | Authors: | Convery, M.A, Young, R.J, Senger, S, Hamblin, J.N, Chan, C, Toomey, J.R, Watson, N.S. | | Deposit date: | 2015-02-13 | | Release date: | 2015-09-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Factor Xa complex with GTC000441
to be published
|
|
