4ASA
 
 | | The structure of modified benzoquinone ansamycins bound to yeast N- terminal Hsp90 | | Descriptor: | ATP-DEPENDENT MOLECULAR CHAPERONE HSP82, CHLORIDE ION, [(3R,5S,6R,7R,12E)-5,11-dimethoxy-3,7,9,15,19-pentamethyl-6-oxidanyl-16,20,22-tris(oxidanylidene)-21-(prop-2-enylamino)-17-azabicyclo[16.3.1]docosa-1(21),8,12,14,18-pentaen-10-yl] carbamate | | Authors: | Roe, S.M, Prodromou, C. | | Deposit date: | 2012-04-30 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Synthesis of 19-Substituted Geldanamycins with Altered Conformations and Their Binding to Heat Shock Protein Hsp90.
Nat.Chem., 5, 2013
|
|
1TFS
 
 | | NMR AND RESTRAINED MOLECULAR DYNAMICS STUDY OF THE THREE-DIMENSIONAL SOLUTION STRUCTURE OF TOXIN FS2, A SPECIFIC BLOCKER OF THE L-TYPE CALCIUM CHANNEL, ISOLATED FROM BLACK MAMBA VENOM | | Descriptor: | TOXIN FS2 | | Authors: | Albrand, J.-P, Blackledge, M.J, Pascaud, F, Hollecker, M, Marion, D. | | Deposit date: | 1995-01-26 | | Release date: | 1995-03-31 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR and restrained molecular dynamics study of the three-dimensional solution structure of toxin FS2, a specific blocker of the L-type calcium channel, isolated from black mamba venom.
Biochemistry, 34, 1995
|
|
7O3C
 
 | | Murine supercomplex CIII2CIV in the mature unlocked conformation | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | Authors: | Vercellino, I, Sazanov, L.A. | | Deposit date: | 2021-04-01 | | Release date: | 2021-10-13 | | Last modified: | 2021-10-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure and assembly of the mammalian mitochondrial supercomplex CIII 2 CIV.
Nature, 598, 2021
|
|
7O37
 
 | | Murine supercomplex CIII2CIV in the assembled locked conformation | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | Authors: | Vercellino, I, Sazanov, L.A. | | Deposit date: | 2021-04-01 | | Release date: | 2021-10-13 | | Last modified: | 2021-10-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure and assembly of the mammalian mitochondrial supercomplex CIII 2 CIV.
Nature, 598, 2021
|
|
7O3E
 
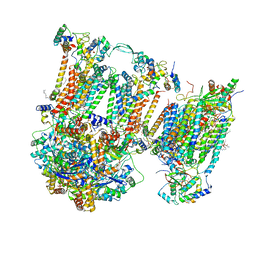 | | Murine supercomplex CIII2CIV in the intermediate locked conformation | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | Authors: | Vercellino, I, Sazanov, L.A. | | Deposit date: | 2021-04-01 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and assembly of the mammalian mitochondrial supercomplex CIII 2 CIV.
Nature, 598, 2021
|
|
5KY8
 
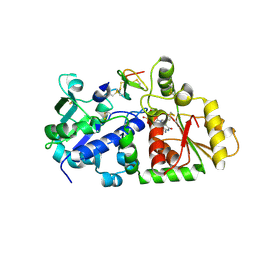 | |
5L0R
 
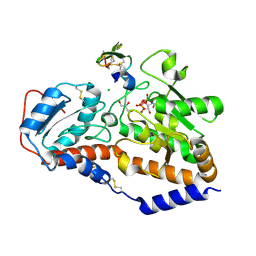 | | human POGLUT1 in complex with Notch1 EGF12 and UDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-09 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of Notch O-glucosylation and O-xylosylation by mammalian protein-O-glucosyltransferase 1 (POGLUT1).
Nat Commun, 8, 2017
|
|
1ZWH
 
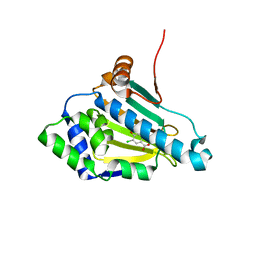 | |
5UB5
 
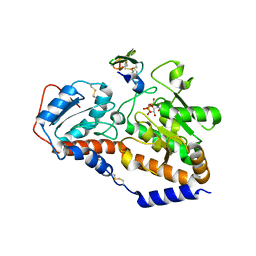 | | human POGLUT1 in complex with human Notch1 EGF12 S458T mutant and UDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neurogenic locus notch homolog protein 1, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-12-20 | | Release date: | 2017-08-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.089 Å) | | Cite: | Structural basis of Notch O-glucosylation and O-xylosylation by mammalian protein-O-glucosyltransferase 1 (POGLUT1).
Nat Commun, 8, 2017
|
|
2YGA
 
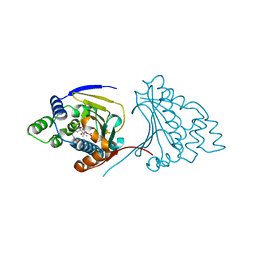 | | E88G-N92L Mutant of N-Term HSP90 complexed with Geldanamycin | | Descriptor: | ATP-DEPENDENT MOLECULAR CHAPERONE HSP82, GELDANAMYCIN | | Authors: | Roe, S.M, Prodromou, C, Pearl, L.H. | | Deposit date: | 2011-04-12 | | Release date: | 2011-11-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Features of the Streptomyces Hygroscopicus Htpg Reveal How Partial Geldanamycin Resistance Can Arise with Mutation to the ATP Binding Pocket of a Eukaryotic Hsp90.
Faseb J., 25, 2011
|
|
8CHY
 
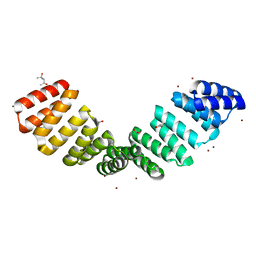 | | Crystal structure of an 8-repeat consensus TPR superhelix with Zinc. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Liutkus, M, Rojas, A.L, Cortajarena, A.L. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Diverse crystalline protein scaffolds through metal-dependent polymorphism.
Protein Sci., 33, 2024
|
|
8CH0
 
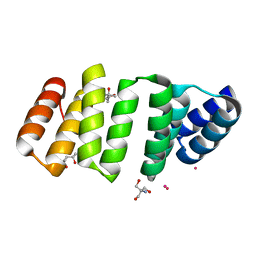 | | Crystal structure of an 8-repeat consensus TPR superhelix with Gadolinium. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Consensus tetratricopeptide repeat protein, ... | | Authors: | Liutkus, M, Rojas, A.L, Cortajarena, A.L. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Diverse crystalline protein scaffolds through metal-dependent polymorphism.
Protein Sci., 33, 2024
|
|
8CIG
 
 | | Crystal structure of an 8-repeat consensus TPR superhelix in tris Buffer with Calcium. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Liutkus, M, Rojas, A.L, Cortajarena, A.L. | | Deposit date: | 2023-02-09 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Diverse crystalline protein scaffolds through metal-dependent polymorphism.
Protein Sci., 33, 2024
|
|
6CGX
 
 | | Backbone cyclised conotoxin Vc1.1 mutant - D11A, E14A | | Descriptor: | Alpha-conotoxin Vc1A | | Authors: | Clark, R.J. | | Deposit date: | 2018-02-21 | | Release date: | 2018-05-23 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure-Activity Studies Reveal the Molecular Basis for GABAB-Receptor Mediated Inhibition of High Voltage-Activated Calcium Channels by alpha-Conotoxin Vc1.1.
ACS Chem. Biol., 13, 2018
|
|
1EZR
 
 | | CRYSTAL STRUCTURE OF NUCLEOSIDE HYDROLASE FROM LEISHMANIA MAJOR | | Descriptor: | CALCIUM ION, NUCLEOSIDE HYDROLASE | | Authors: | Shi, W, Schramm, V.L, Almo, S.C. | | Deposit date: | 2000-05-11 | | Release date: | 2000-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nucleoside hydrolase from Leishmania major. Cloning, expression, catalytic properties, transition state inhibitors, and the 2.5-a crystal structure.
J.Biol.Chem., 274, 1999
|
|
4QHR
 
 | | The structure of alanine racemase from Acinetobacter baumannii | | Descriptor: | Alanine racemase | | Authors: | Davis, E, Scaletti-Hutchinson, E, Nakatani, Y, Krause, K.L. | | Deposit date: | 2014-05-29 | | Release date: | 2015-05-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of alanine racemase from Acinetobacter baumannii
ACTA CRYSTALLOGR.,SECT.F, 70, 2014
|
|
7ZOR
 
 | | Crystal structure of anti-Siglec-15 Fab | | Descriptor: | Anti-siglec15 FAB HC, Anti-siglec15 FAB LC | | Authors: | Lenza, M.P, Oyenarte, I, Jimenez-Barbero, J, Ereno-Orbea, J. | | Deposit date: | 2022-04-26 | | Release date: | 2023-06-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.933 Å) | | Cite: | Structural insights into Siglec-15 reveal glycosylation dependency for its interaction with T cells through integrin CD11b.
Nat Commun, 14, 2023
|
|
7ZOZ
 
 | | Crystal structure of Siglec-15 in complex with Fab | | Descriptor: | Anti-Siglec 15 Fab HC, Anti-Siglec 15 Fab LC, Sialic acid-binding Ig-like lectin 15 | | Authors: | Lenza, M.P, Oyenarte, I, Jimenez-Barbero, J, Ereno-Orbea, J. | | Deposit date: | 2022-04-26 | | Release date: | 2023-06-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Structural insights into Siglec-15 reveal glycosylation dependency for its interaction with T cells through integrin CD11b.
Nat Commun, 14, 2023
|
|
7ZZ5
 
 | | Cryo-EM structure of "BC open" conformation of Lactococcus lactis pyruvate carboxylase with acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, BICARBONATE ION, Pyruvate carboxylase | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
7ZZ3
 
 | | Cryo-EM structure of "BC react" conformation of Lactococcus lactis pyruvate carboxylase with acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
7ZYZ
 
 | | Cryo-EM structure of "CT oxa" conformation of Lactococcus lactis pyruvate carboxylase with acetyl-CoA | | Descriptor: | MANGANESE (II) ION, OXALOACETATE ION, Pyruvate carboxylase | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-10-12 | | Last modified: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
7ZZ1
 
 | | Cryo-EM structure of "CT react" conformation of Lactococcus lactis pyruvate carboxylase with acetyl-CoA | | Descriptor: | BIOTIN, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-10-12 | | Last modified: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
7ZZ2
 
 | | Cryo-EM structure of "CT pyr" conformation of Lactococcus lactis pyruvate carboxylase with acetyl-CoA | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, PYRUVIC ACID, ... | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
7ZZ0
 
 | | Cryo-EM structure of "CT empty" conformation of Lactococcus lactis pyruvate carboxylase with acetyl-CoA | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, Pyruvate carboxylase | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-10-12 | | Last modified: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
7ZZ4
 
 | | Cryo-EM structure of "BC closed" conformation of Lactococcus lactis pyruvate carboxylase with acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
