4USZ
 
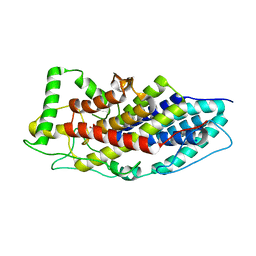 | | Crystal structure of the first bacterial vanadium dependant iodoperoxidase | | Descriptor: | SODIUM ION, VANADATE ION, VANADIUM-DEPENDENT HALOPEROXIDASE | | Authors: | Rebuffet, E, Delage, L, Fournier, J.B, Rzonca, J, Potin, P, Michel, G, Czjzek, M, Leblanc, C. | | Deposit date: | 2014-07-17 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Bacterial Vanadium Iodoperoxidase from the Marine Flavobacteriaceae Zobellia Galactanivorans Reveals Novel Molecular and Evolutionary Features of Halide Specificity in This Enzyme Family.
Appl.Environ.Microbiol., 80, 2014
|
|
4USW
 
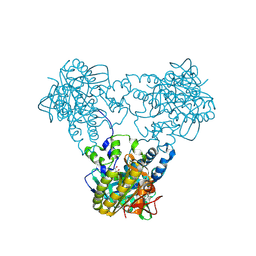 | | Crystal structure of human soluble Adenylyl Cyclase with ATP | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, ADENYLATE CYCLASE TYPE 10, ... | | Authors: | Kleinboelting, S, Steegborn, C. | | Deposit date: | 2014-07-13 | | Release date: | 2014-07-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Analysis of Human Soluble Adenylyl Cyclase and Crystal Structures of its Nucleotide Complexes -Implications for Cyclase Catalysis and Evolution.
FEBS J., 281, 2014
|
|
4V16
 
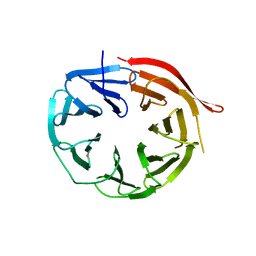 | | KlHsv2 with loop 6CD replaced by a Gly-Ser linker | | Descriptor: | SVP1-LIKE PROTEIN 2 | | Authors: | Busse, R.A, Scacioc, A, Krick, R, Perez-Lara, A, Thumm, M, Kuhnel, K. | | Deposit date: | 2014-09-25 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Characterization of Proppin-Phosphoinositide Binding and Role of Loop 6Cd in Proppin-Membrane Binding.
Biophys.J., 108, 2015
|
|
4UQS
 
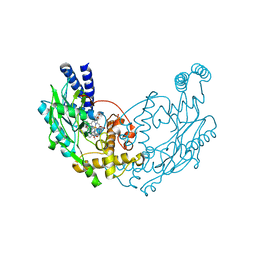 | |
4UQV
 
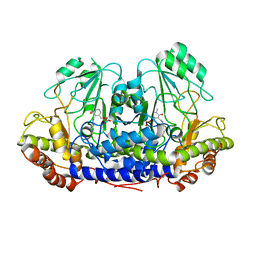 | |
4TWB
 
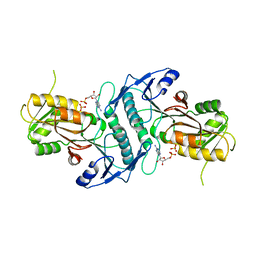 | | Sulfolobus solfataricus ribose-phosphate pyrophosphokinase | | Descriptor: | ADENOSINE MONOPHOSPHATE, Ribose-phosphate pyrophosphokinase, SULFATE ION | | Authors: | Kadziola, A. | | Deposit date: | 2014-06-30 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structure of dimeric, recombinant Sulfolobus solfataricus phosphoribosyl diphosphate synthase: a bent dimer defining the adenine specificity of the substrate ATP.
Extremophiles, 19, 2015
|
|
4UXD
 
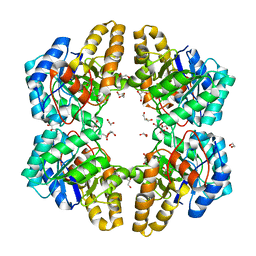 | | 2-keto 3-deoxygluconate aldolase from Picrophilus torridus | | Descriptor: | 1,2-ETHANEDIOL, 2-DEHYDRO-3-DEOXY-D-GLUCONATE/2-DEHYDRO-3-DEOXY-PHOSPHOGLUCONATE ALDOLASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Priftis, A, Zaitsev, V, Reher, M, Johnsen, U, Danson, M.J, Taylor, G.L, Schoenheit, P, Crennell, S.J. | | Deposit date: | 2014-08-22 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into the Substrate Specificity of Archaeal Entner-Doudoroff Aldolases: The Structures of Picrophilus torridus 2-Keto-3-deoxygluconate Aldolase and Sulfolobus solfataricus 2-Keto-3-deoxy-6-phosphogluconate Aldolase in Complex with 2-Keto-3-deoxy-6-phosphogluconate.
Biochemistry, 57, 2018
|
|
5OW7
 
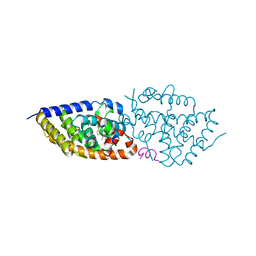 | | VDR complex | | Descriptor: | (3~{S})-3-[(1~{S},3~{a}~{S},4~{E},7~{a}~{S})-7~{a}-methyl-4-[(2~{Z})-2-[(5~{S})-2-methylidene-5-oxidanyl-cyclohexylidene]ethylidene]-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-1-yl]-3-oxidanyl-~{N}-propan-2-yl-butanamide, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Rochel, N, Li, W. | | Deposit date: | 2017-08-31 | | Release date: | 2018-02-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Investigation of 20S-hydroxyvitamin D3 analogs and their 1 alpha-OH derivatives as potent vitamin D receptor agonists with anti-inflammatory activities.
Sci Rep, 8, 2018
|
|
4U2C
 
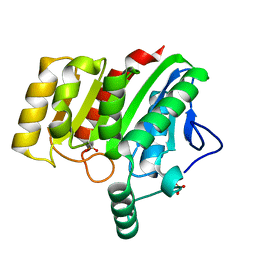 | | Crystal structure of dienelactone hydrolase A-6 variant (S7T, A24V, Q35H, F38L, Q110L, C123S, Y145C, E199G and S208G) at 1.95 A resolution | | Descriptor: | Carboxymethylenebutenolidase, SULFATE ION | | Authors: | Porter, J.L, Collyer, C.A, Ollis, D.L. | | Deposit date: | 2014-07-16 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Directed evolution of new and improved enzyme functions using an evolutionary intermediate and multidirectional search.
Acs Chem.Biol., 10, 2015
|
|
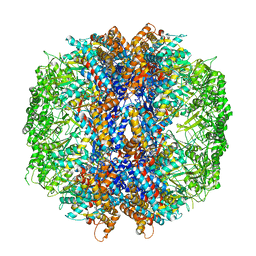
4V4K
 
 | |
4U2D
 
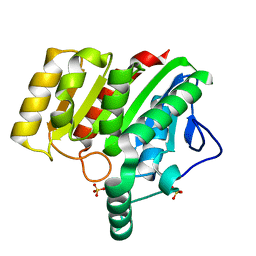 | | Crystal structure of dienelactone hydrolase S-2 variant (Q35H, F38L, Q110L, C123S, Y137C, Y145C, N154D, E199G, S208G and G211D) at 1.67 A resolution | | Descriptor: | Carboxymethylenebutenolidase, SULFATE ION | | Authors: | Porter, J.L, Collyer, C.A, Ollis, D.L. | | Deposit date: | 2014-07-16 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Directed evolution of new and improved enzyme functions using an evolutionary intermediate and multidirectional search.
Acs Chem.Biol., 10, 2015
|
|
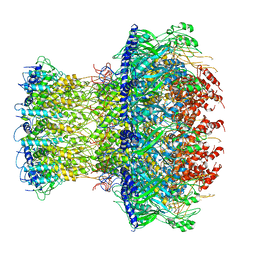
4V8R
 
 | | The crystal structures of the eukaryotic chaperonin CCT reveal its functional partitioning | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Kalisman, N, Schroder, G.F, Levitt, M. | | Deposit date: | 2012-03-28 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The Crystal Structures of the Eukaryotic Chaperonin Cct Reveal its Functional Partitioning
Structure, 21, 2013
|
|
4U2G
 
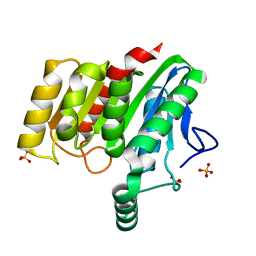 | | Crystal structure of dienelactone hydrolase B-4 variant (Q35H, F38L, Y64H, Q76L, Q110L, C123S, Y137C, A141V, Y145C, N154D, E199G, S208G, G211D, S233G and 237Q) at 1.80 A resolution | | Descriptor: | Carboxymethylenebutenolidase, SULFATE ION | | Authors: | Porter, J.L, Collyer, C.A, Ollis, D.L. | | Deposit date: | 2014-07-16 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Directed evolution of new and improved enzyme functions using an evolutionary intermediate and multidirectional search.
Acs Chem.Biol., 10, 2015
|
|
4TUH
 
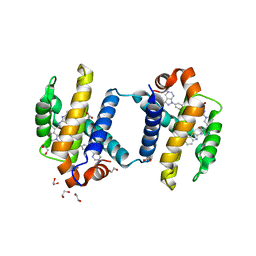 | | Bcl-xL in complex with inhibitor (Compound 10) | | Descriptor: | 1,2-ETHANEDIOL, 2-[8-(1,3-benzothiazol-2-ylcarbamoyl)-3,4-dihydroisoquinolin-2(1H)-yl]-5-{3-[4-(1H-pyrazolo[3,4-d]pyrimidin-1-yl)phenoxy]propyl}-1,3-thiazole-4-carboxylic acid, ACETATE ION, ... | | Authors: | Czabotar, P.E, Lessense, G, Smith, B.J, Colman, P.M. | | Deposit date: | 2014-06-24 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Guided Rescaffolding of Selective Antagonists of BCL-XL.
Acs Med.Chem.Lett., 5, 2014
|
|
4UTK
 
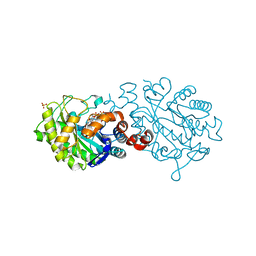 | | XenA - reduced - Y183F variant | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, SULFATE ION, XENOBIOTIC REDUCTASE | | Authors: | Werther, T, Dobbek, H. | | Deposit date: | 2014-07-21 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Redox-dependent substrate-cofactor interactions in the Michaelis-complex of a flavin-dependent oxidoreductase
Nat Commun, 8, 2017
|
|
4V08
 
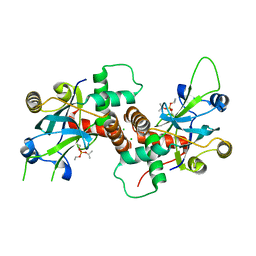 | | Inhibited dimeric pseudorabies virus protease pUL26N at 2 A resolution | | Descriptor: | CHLORIDE ION, DIISOPROPYL PHOSPHONATE, MAGNESIUM ION, ... | | Authors: | Zuehlsdorf, M, Werten, S, Palm, G.J, Hinrichs, W. | | Deposit date: | 2014-09-11 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Dimerization-Induced Allosteric Changes of the Oxyanion-Hole Loop Activate the Pseudorabies Virus Assemblin Pul26N, a Herpesvirus Serine Protease
Plos Pathog., 11, 2015
|
|
5OYH
 
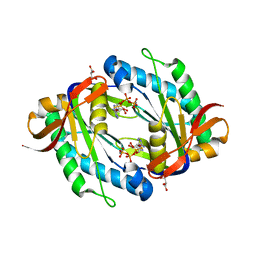 | | crystal structure of the catalytic core of a rhodopsin-guanylyl cyclase with converted specificity in complex with ATPalphaS | | Descriptor: | ADENOSINE-5'-SP-ALPHA-THIO-TRIPHOSPHATE, CALCIUM ION, GLYCEROL, ... | | Authors: | Broser, M, Scheib, U, Hegemann, P. | | Deposit date: | 2017-09-09 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | Rhodopsin-cyclases for photocontrol of cGMP/cAMP and 2.3 angstrom structure of the adenylyl cyclase domain.
Nat Commun, 9, 2018
|
|
4UV2
 
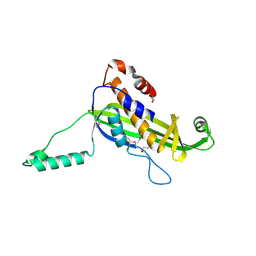 | | Structure of the curli transport lipoprotein CsgG in a non-lipidated, pre-pore conformation | | Descriptor: | CURLI PRODUCTION TRANSPORT COMPONENT CSGG | | Authors: | Goyal, P, Krasteva, P.V, Gerven, N.V, Gubellini, F, Broeck, I.V.D, Troupiotis-Tsailaki, A, Jonckheere, W, Pehau-Arnaudet, G, Pinkner, J.S, Chapman, M.R, Hultgren, S.J, Howorka, S, Fronzes, R, Remaut, H. | | Deposit date: | 2014-08-04 | | Release date: | 2014-09-24 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Mechanistic Insights Into the Bacterial Amyloid Secretion Channel Csgg.
Nature, 516, 2014
|
|
4U7Q
 
 | | Structure of wild-type HIV protease in complex with photosensitive inhibitor PDI-6 | | Descriptor: | N~2~-({[7-(diethylamino)-2-oxo-2H-chromen-4-yl]methoxy}carbonyl)-N-[(2S,4S,5S)-4-hydroxy-1,6-diphenyl-5-{[(1,3-thiazol-5-ylmethoxy)carbonyl]amino}hexan-2-yl]-L-valinamide, V-1 protease | | Authors: | Pachl, P, Rezacova, P, Schimer, J. | | Deposit date: | 2014-07-31 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Triggering HIV polyprotein processing by light using rapid photodegradation of a tight-binding protease inhibitor.
Nat Commun, 6, 2015
|
|
4U7V
 
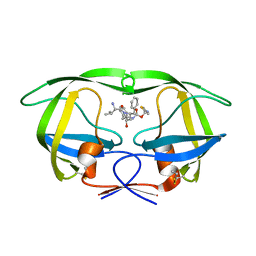 | | Structure of wild-type HIV protease in complex with degraded photosensitive inhibitor | | Descriptor: | BETA-MERCAPTOETHANOL, N-[(2S,4S,5S)-4-hydroxy-1,6-diphenyl-5-{[(1,3-thiazol-5-ylmethoxy)carbonyl]amino}hexan-2-yl]-L-valinamide, V-1 protease | | Authors: | Pachl, P, Rezacova, P, Schimer, J. | | Deposit date: | 2014-07-31 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Triggering HIV polyprotein processing by light using rapid photodegradation of a tight-binding protease inhibitor.
Nat Commun, 6, 2015
|
|
4V2T
 
 | | Membrane embedded pleurotolysin pore with 13 fold symmetry | | Descriptor: | PLEUROTOLYSIN A, PLEUROTOLYSIN B | | Authors: | Lukoyanova, N, Kondos, S.C, Farabella, I, Law, R.H.P, Reboul, C.F, Caradoc-Davies, T.T, Spicer, B.A, Kleifeld, O, Perugini, M, Ekkel, S, Hatfaludi, T, Oliver, K, Hotze, E.M, Tweten, R.K, Whisstock, J.C, Topf, M, Dunstone, M.A, Saibil, H.R. | | Deposit date: | 2014-10-15 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Conformational Changes During Pore Formation by the Perforin-Related Protein Pleurotolysin.
Plos Biol., 13, 2015
|
|
4V41
 
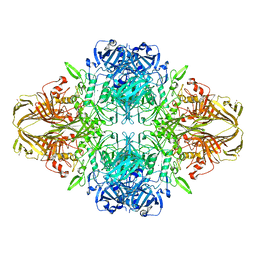 | | E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) | | Descriptor: | BETA-GALACTOSIDASE, MAGNESIUM ION | | Authors: | Juers, D.H, Jacobson, R.H, Wigley, D, Zhang, X.J, Huber, R.E, Tronrud, D.E, Matthews, B.W. | | Deposit date: | 2000-06-07 | | Release date: | 2014-07-09 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation.
Protein Sci., 9, 2000
|
|
4U8Z
 
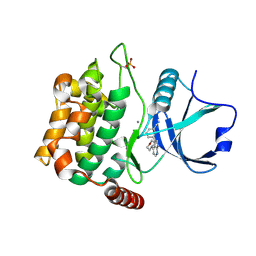 | | Crystal structure of MST3 with a pyrrolopyrimidine inhibitor (PF-06447475) | | Descriptor: | 3-[4-(morpholin-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile, MANGANESE (II) ION, Serine/threonine-protein kinase 24 | | Authors: | Jasti, J, Song, X, Griffor, M, Kurumbail, R.G. | | Deposit date: | 2014-08-05 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Discovery and preclinical profiling of 3-[4-(morpholin-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile (PF-06447475), a highly potent, selective, brain penetrant, and in vivo active LRRK2 kinase inhibitor.
J.Med.Chem., 58, 2015
|
|
4V5W
 
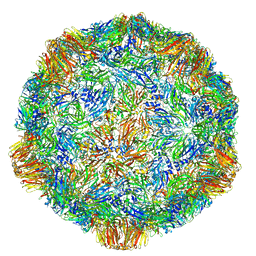 | | Grapevine Fanleaf virus | | Descriptor: | COAT PROTEIN | | Authors: | Schellenberger, P, Demangeat, G, Ritzenthaler, C, Lorber, B, Sauter, C. | | Deposit date: | 2011-05-10 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Exploiting Protein Engineering and Crystal Polymorphism for Successful X-Ray Structure Determination
Cryst.Growth Des., 11, 2011
|
|
4W9E
 
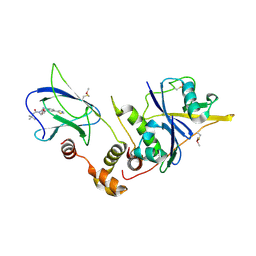 | | pVHL:EloB:EloC in complex with (2S,4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(thiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 4) | | Descriptor: | (4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-[4-(1,3-thiazol-5-yl)benzyl]-L-prolinamide, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | Authors: | Gadd, M.S, Soares, P, Galdeano, C, van Molle, I, Ciulli, A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Guided Design and Optimization of Small Molecules Targeting the Protein-Protein Interaction between the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase and the Hypoxia Inducible Factor (HIF) Alpha Subunit with in Vitro Nanomolar Affinities.
J.Med.Chem., 57, 2014
|
|
