6H3B
 
 | |
5UMJ
 
 | |
6I00
 
 | |
2AUO
 
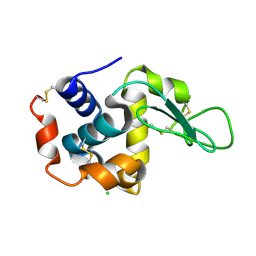
 | | Residue F4 plays a key role in modulating the oxygen affinity and cooperatrivity in Scapharca dimeric hemoglobin | | Descriptor: | CARBON MONOXIDE, Globin I, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Knapp, J.E, Bonham, M.A, Gibson, Q.H, Nichols, J.C, Royer Jr, W.E. | | Deposit date: | 2005-08-28 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Residue F4 plays a key role in modulating oxygen affinity and cooperativity in Scapharca dimeric hemoglobin
Biochemistry, 44, 2005
|
|
7L2Z
 
 | |
1EJC
 
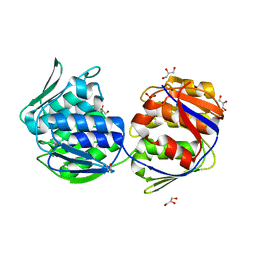 | | Crystal structure of unliganded mura (type2) | | Descriptor: | GLYCEROL, PHOSPHATE ION, UDP-N-ACETYLGLUCOSAMINE ENOLPYRUVYLTRANSFERASE | | Authors: | Eschenburg, S, Schonbrunn, E. | | Deposit date: | 2000-03-02 | | Release date: | 2000-10-25 | | Last modified: | 2011-07-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparative X-ray analysis of the un-liganded fosfomycin-target murA.
Proteins, 40, 2000
|
|
1HLU
 
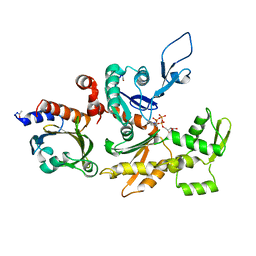 | | STRUCTURE OF BOVINE BETA-ACTIN-PROFILIN COMPLEX WITH ACTIN BOUND ATP PHOSPHATES SOLVENT ACCESSIBLE | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, BETA-ACTIN, CALCIUM ION, ... | | Authors: | Chik, J.K, Lindberg, U, Schutt, C.E. | | Deposit date: | 1997-05-30 | | Release date: | 1997-10-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The structure of an open state of beta-actin at 2.65 A resolution.
J.Mol.Biol., 263, 1996
|
|
6I4K
 
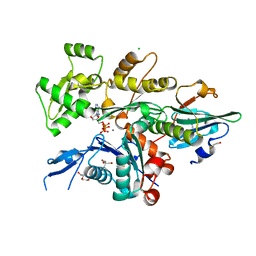 | | Crystal Structure of Plasmodium falciparum actin I (G115A mutant) in the Ca-ATP state | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-TRIPHOSPHATE, Actin-1, ... | | Authors: | Kumpula, E.-P, Lopez, A.J, Tajedin, L, Han, H, Kursula, I. | | Deposit date: | 2018-11-09 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Atomic view into Plasmodium actin polymerization, ATP hydrolysis, and fragmentation.
Plos Biol., 17, 2019
|
|
7KWU
 
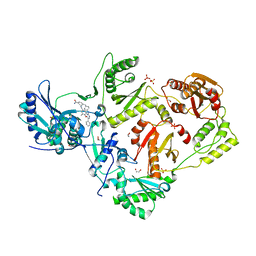 | | Crystal Structure of HIV-1 RT in Complex with 16c (K07-15) | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-{[4-(4-cyano-2,6-dimethylphenoxy)-5-(pyridin-4-yl)pyrimidin-2-yl]amino}piperidin-1-yl)methyl]benzamide, MAGNESIUM ION, ... | | Authors: | Ruiz, F.X, Arnold, E. | | Deposit date: | 2020-12-02 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | 2,4,5-Trisubstituted Pyrimidines as Potent HIV-1 NNRTIs: Rational Design, Synthesis, Activity Evaluation, and Crystallographic Studies.
J.Med.Chem., 64, 2021
|
|
1HP7
 
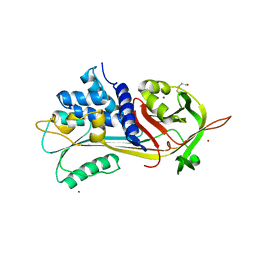 | | A 2.1 ANGSTROM STRUCTURE OF AN UNCLEAVED ALPHA-1-ANTITRYPSIN SHOWS VARIABILITY OF THE REACTIVE CENTER AND OTHER LOOPS | | Descriptor: | ALPHA-1-ANTITRYPSIN, BETA-MERCAPTOETHANOL, ZINC ION | | Authors: | Kim, S.-J, Woo, J.-R, Seo, E.J, Yu, M.-H, Ryu, S.-E. | | Deposit date: | 2000-12-12 | | Release date: | 2001-03-14 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A 2.1 A resolution structure of an uncleaved alpha(1)-antitrypsin shows variability of the reactive center and other loops.
J.Mol.Biol., 306, 2001
|
|
4CEV
 
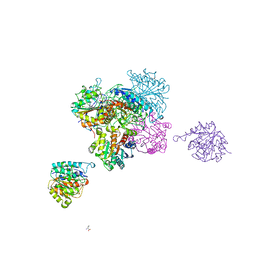 | | ARGINASE FROM BACILLUS CALDEVELOX, L-ORNITHINE COMPLEX | | Descriptor: | GUANIDINE, L-ornithine, MANGANESE (II) ION, ... | | Authors: | Bewley, M.C, Jeffrey, P.D, Patchett, M.L, Kanyo, Z.F, Baker, E.N. | | Deposit date: | 1999-03-15 | | Release date: | 1999-04-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of Bacillus caldovelox arginase in complex with substrate and inhibitors reveal new insights into activation, inhibition and catalysis in the arginase superfamily.
Structure Fold.Des., 7, 1999
|
|
4BPA
 
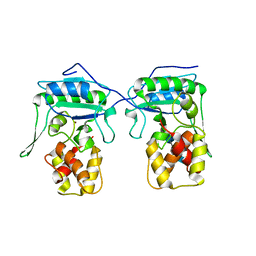 | | Crystal structure of AmpDh2 from Pseudomonas aeruginosa in complex with NAG-NAM-NAG-NAM tetrasaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-beta-D-glucopyranoside, AMPDH2, ZINC ION | | Authors: | Artola-Recolons, C, Martinez-Caballero, S, Lee, M, Carrasco-Lopez, C, Hesek, D, Spink, E, Lastochkin, E, Zhang, W, Hellman, L, Boggess, B, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2013-05-23 | | Release date: | 2013-07-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Reaction Products and the X-Ray Structure of Ampdh2, a Virulence Determinant of Pseudomonas Aeruginosa.
J.Am.Chem.Soc., 135, 2013
|
|
6GNM
 
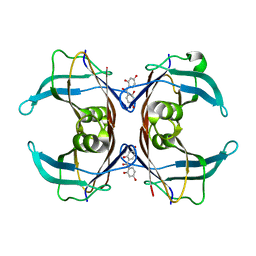 | | Crystal Structure Of Sea Bream Transthyretin in complex with 2,2',4,4'-tetrahydroxybenzophenone (BP2) | | Descriptor: | Transthyretin, bis(2,4-dihydroxyphenyl)methanone | | Authors: | Grundstrom, C, Zhang, J, Olofsson, A, Andersson, P.L, Sauer-Eriksson, A.E. | | Deposit date: | 2018-05-31 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Interspecies Variation between Fish and Human Transthyretins in Their Binding of Thyroid-Disrupting Chemicals.
Environ. Sci. Technol., 52, 2018
|
|
2X9I
 
 | |
6GNR
 
 | | Crystal Structure Of Sea Bream Transthyretin in complex with 2-(3-chloro-2-methylanilino)pyridine-3-carboxylic acid (Clonixin) | | Descriptor: | 2-(3-chloro-2-methylanilino)pyridine-3-carboxylic acid, Transthyretin | | Authors: | Grundstrom, C, Zhang, J, Olofsson, A, Andersson, P.L, Sauer-Eriksson, A.E. | | Deposit date: | 2018-05-31 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Interspecies Variation between Fish and Human Transthyretins in Their Binding of Thyroid-Disrupting Chemicals.
Environ. Sci. Technol., 52, 2018
|
|
6GWR
 
 | | Structure of the kinase domain of human DDR1 in complex with a potent and selective inhibitor of DDR1 and DDR2 | | Descriptor: | 1,2-ETHANEDIOL, 3-(2-imidazo[1,2-a]pyrazin-3-ylethynyl)-~{N}-[3-[(4-methylpiperazin-1-yl)methyl]-5-(trifluoromethyl)phenyl]-4-propan-2-yl-benzamide, Epithelial discoidin domain-containing receptor 1, ... | | Authors: | Pinkas, D.M, Fox, A.E, Kupinska, K, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2018-06-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 3-(Imidazo[1,2- a]pyrazin-3-ylethynyl)-4-isopropyl- N-(3-((4-methylpiperazin-1-yl)methyl)-5-(trifluoromethyl)phenyl)benzamide as a Dual Inhibitor of Discoidin Domain Receptors 1 and 2.
J. Med. Chem., 61, 2018
|
|
4PII
 
 | | Crystal structure of hypothetical protein PF0907 from pyrococcus furiosus solved by sulfur SAD using Swiss light source data | | Descriptor: | CHLORIDE ION, IMIDAZOLE, N-glycosylase/DNA lyase | | Authors: | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2014-05-08 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
3Q8M
 
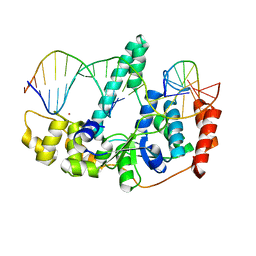 | | Crystal Structure of Human Flap Endonuclease FEN1 (D181A) in complex with substrate 5'-flap DNA and K+ | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*AP*CP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*T)-3'), DNA (5'-D(*TP*TP*GP*AP*GP*GP*CP*AP*GP*AP*GP*T)-3'), ... | | Authors: | Tsutakawa, S.E, Classen, S, Chapados, B.R, Arvai, A, Finger, D.L, Guenther, G, Tomlinson, C.G, Thompson, P, Sarker, A.H, Shen, B, Cooper, P.K, Grasby, J.A, Tainer, J.A. | | Deposit date: | 2011-01-06 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human Flap Endonuclease Structures, DNA Double-Base Flipping, and a Unified Understanding of the FEN1 Superfamily.
Cell(Cambridge,Mass.), 145, 2011
|
|
2WWN
 
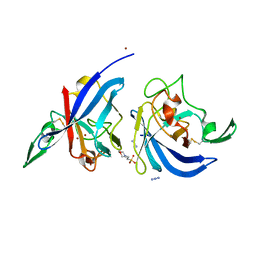 | | Yersinia pseudotuberculosis Superoxide Dismutase C with bound Azide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, AZIDE ION, SUPEROXIDE DISMUTASE [CU-ZN], ... | | Authors: | Basak, A.K, Duffield, M.L, Naylor, C.E, Huyet, J, Titball, R.W. | | Deposit date: | 2009-10-26 | | Release date: | 2010-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Yersinia Pseudotuberculosis Superoxide Dismutase (Sodc)
To be Published
|
|
3EIX
 
 | |
4JBF
 
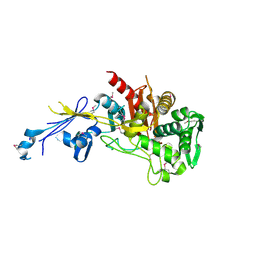 | | Crystal structure of peptidoglycan glycosyltransferase from Atopobium parvulum DSM 20469. | | Descriptor: | Peptidoglycan glycosyltransferase, TETRAETHYLENE GLYCOL | | Authors: | Filippova, E.V, Wawrzak, Z, Minasov, G, Shuvalova, L, Kiryukhina, O, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2013-02-19 | | Release date: | 2013-03-20 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of peptidoglycan glycosyltransferase from Atopobium parvulum DSM 20469.
To be Published
|
|
2X7S
 
 | | Structures of human carbonic anhydrase II inhibitor complexes reveal a second binding site for steroidal and non-steroidal inhibitors. | | Descriptor: | (13ALPHA,14BETA,17ALPHA)-3-HYDROXY-2-METHOXYESTRA-1,3,5(10)-TRIEN-17-YL SULFAMATE, CARBONIC ANHYDRASE 2, GLYCEROL, ... | | Authors: | Cozier, G.E, Leese, M.P, Lloyd, M.D, Baker, M.D, Thiyagarajan, N, Acharya, K.R, Potter, B.V.L. | | Deposit date: | 2010-03-03 | | Release date: | 2010-03-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structures of Human Carbonic Anhydrase II/Inhibitor Complexes Reveal a Second Binding Site for Steroidal and Nonsteroidal Inhibitors.
Biochemistry, 49, 2010
|
|
6GR7
 
 | | Crystal Structure Of Human Transthyretin in complex with 2,4,5-trichlorophenoxyacetic acid (2,4,5-T) | | Descriptor: | 2-[2,4,5-tris(chloranyl)phenoxy]ethanoic acid, Transthyretin | | Authors: | Grundstrom, C, Zhang, J, Olofsson, A, Andersson, P.L, Sauer-Eriksson, A.E. | | Deposit date: | 2018-06-09 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Interspecies Variation between Fish and Human Transthyretins in Their Binding of Thyroid-Disrupting Chemicals.
Environ. Sci. Technol., 52, 2018
|
|
2WOO
 
 | | Nucleotide-free form of S. pombe Get3 | | Descriptor: | ATPASE GET3, ZINC ION | | Authors: | Mateja, A, Szlachcic, A, Downing, M.E, Dobosz, M, Mariappan, M, Hegde, R.S, Keenan, R.J. | | Deposit date: | 2009-07-27 | | Release date: | 2009-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.006 Å) | | Cite: | The Structural Basis of Tail-Anchored Membrane Protein Recognition by Get3.
Nature, 461, 2009
|
|
2CYD
 
 | | Crystal structure of Lithium bound rotor ring of the V-ATPase from Enterococcus hirae | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, LITHIUM ION, UNDECYL-MALTOSIDE, ... | | Authors: | Murata, T, Yamato, I, Kakinuma, Y, Shirouzu, M, Walker, J.E, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-06 | | Release date: | 2006-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Lithium bound rotor ring of the V-ATPase from Enterococcus hirae
To be Published
|
|
