7OER
 
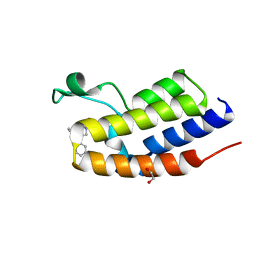 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH N-(2,2-diphenylethyl)-1,5-dimethyl-N-(2-(methylamino)-2-oxoethyl)-6-oxo-1,6-dihydropyridine-3-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 2, N-(2,2-diphenylethyl)-1,5-dimethyl-N-[2-(methylamino)-2-oxidanylidene-ethyl]-6-oxidanylidene-pyridine-3-carboxamide | | Authors: | Chung, C. | | Deposit date: | 2021-05-03 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Discovery of a Highly Selective BET BD2 Inhibitor from a DNA-Encoded Library Technology Screening Hit.
J.Med.Chem., 64, 2021
|
|
4UWS
 
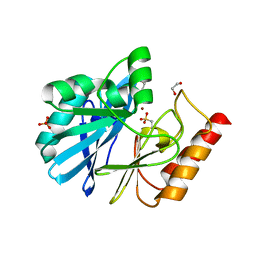 | | VIM-26-PEG. Leu224 in VIM-26 from Klebsiella pneumoniae has implications for drug binding. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, METALLO-BETA-LACTAMASE VIM-26, ... | | Authors: | Leiros, H.-K.S, Edvardsen, K.S.W, Bjerga, G.E.K, Samuelsen, O. | | Deposit date: | 2014-08-14 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural and Biochemical Characterization of Vim-26 Show that Leu224 Has Implications for the Substrate Specificity of Vim Metallo-Beta-Lactamases.
FEBS J., 282, 2015
|
|
8GBW
 
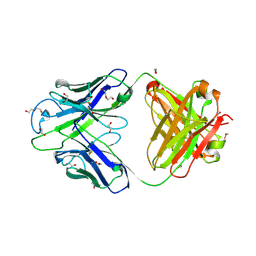 | | Crystal structure of PC39-23D, an anti-HIV broadly neutralizing antibody | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, PC39-23D Fab heavy chain, ... | | Authors: | Murrell, S, Omorodion, O, Wilson, I.A. | | Deposit date: | 2023-02-28 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Antigen pressure from two founder viruses induces multiple insertions at a single antibody position to generate broadly neutralizing HIV antibodies.
Plos Pathog., 19, 2023
|
|
8GBV
 
 | | Crystal structure of PC39-17A, an anti-HIV broadly neutralizing antibody | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, PC39-17A Fab heavy chain, ... | | Authors: | Murrell, S, Omorodion, O, Wilson, I.A. | | Deposit date: | 2023-02-28 | | Release date: | 2023-06-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Antigen pressure from two founder viruses induces multiple insertions at a single antibody position to generate broadly neutralizing HIV antibodies.
Plos Pathog., 19, 2023
|
|
7L6Z
 
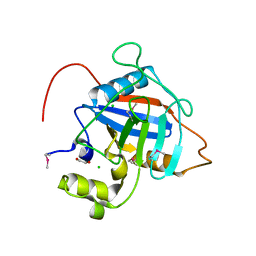 | | Crystal Structure of Peptidyl-Prolyl Cis-Trans Isomerasefrom (PpiB) Streptococcus pneumoniae R6 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-24 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of Peptidyl-Prolyl Cis-Trans Isomerasefrom (PpiB) Streptococcus pneumoniae R6
To Be Published
|
|
5LV2
 
 | | Crystal structure of mouse CARM1 in complex with inhibitor LH1246 | | Descriptor: | 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, 1-METHOXY-2-(2-METHOXYETHOXY)ETHANE, ... | | Authors: | Cura, V, Marechal, N, Troffer-Charlier, N, Halby, L, Arimondo, P, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-09-12 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Hijacking DNA methyltransferase transition state analogues to produce chemical scaffolds for PRMT inhibitors.
Philos.Trans.R.Soc.Lond.B Biol.Sci., 373, 2018
|
|
7OKC
 
 | | Crystal structure of Escherichia coli LpxA in complex with compound 1 | | Descriptor: | 2-[2-(2-chlorophenyl)sulfanylethanoyl-[[4-(1,2,4-triazol-1-yl)phenyl]methyl]amino]-N-methyl-ethanamide, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, SODIUM ION | | Authors: | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
5LTT
 
 | |
1IYT
 
 | | Solution structure of the Alzheimer's disease amyloid beta-peptide (1-42) | | Descriptor: | Alzheimer's disease amyloid | | Authors: | Crescenzi, O, Tomaselli, S, Guerrini, R, Salvadori, S, D'Ursi, A.M, Temussi, P.A, Picone, D. | | Deposit date: | 2002-09-06 | | Release date: | 2003-02-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Alzheimer amyloid beta-peptide (1-42) in an apolar microenvironment. Similarity with a virus fusion domain.
EUR.J.BIOCHEM., 269, 2002
|
|
6OZK
 
 | |
5LLU
 
 | | Structure of the thermostabilized EAAT1 cryst-II mutant in complex with L-ASP | | Descriptor: | ASPARTIC ACID, Excitatory amino acid transporter 1,Neutral amino acid transporter B(0),Excitatory amino acid transporter 1, SODIUM ION | | Authors: | Canul-Tec, J, Assal, R, Legrand, P, Reyes, N. | | Deposit date: | 2016-07-28 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Structure and allosteric inhibition of excitatory amino acid transporter 1.
Nature, 544, 2017
|
|
8G7Y
 
 | |
4W7P
 
 | | Crystal Structure of ROCK 1 bound to YB-15-QD37 | | Descriptor: | N~1~-[2-(1H-indazol-5-yl)pyrido[3,4-d]pyrimidin-4-yl]-2-methylpropane-1,2-diamine, Rho-associated protein kinase 1 | | Authors: | Sprague, E.R. | | Deposit date: | 2014-08-22 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Novel ROCK inhibitors for the treatment of pulmonary arterial hypertension.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4W9D
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methyloxazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 3) | | Descriptor: | (4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-[4-(4-methyl-1,3-oxazol-5-yl)benzyl]-L-prolinamide, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | Authors: | Gadd, M.S, Hewitt, S, Galdeano, C, van Molle, I, Ciulli, A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Design and Optimization of Small Molecules Targeting the Protein-Protein Interaction between the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase and the Hypoxia Inducible Factor (HIF) Alpha Subunit with in Vitro Nanomolar Affinities.
J.Med.Chem., 57, 2014
|
|
4USV
 
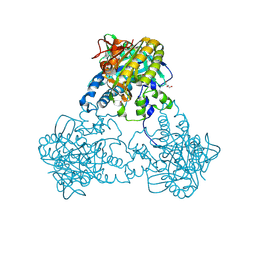 | |
4URL
 
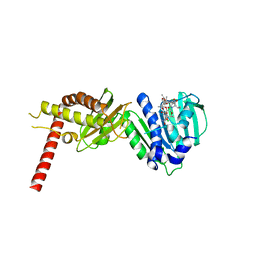 | | Crystal Structure of Staph ParE43kDa in complex with KBD | | Descriptor: | (1R,4aS,5S,6S,8aR)-5-{[(5S)-1-(3-O-acetyl-4-O-carbamoyl-6-deoxy-2-O-methyl-alpha-L-talopyranosyl)-4-hydroxy-2-oxo-5-(propan-2-yl)-2,5-dihydro-1H-pyrrol-3-yl]carbonyl}-6-methyl-4-methylidene-1,2,3,4,4a,5,6,8a-octahydronaphthalen-1-yl 2,6-dideoxy-3-C-[(1S)-1-{[(3,4-dichloro-5-methyl-1H-pyrrol-2-yl)carbonyl]amino}ethyl]-beta-D-ribo-hexopyranoside, DNA TOPOISOMERASE IV, B SUBUNIT | | Authors: | Lu, J, Patel, S, Sharma, N, Soisson, S, Kishii, R, Takei, M, Fukuda, Y, Lumb, K.J, Singh, S.B. | | Deposit date: | 2014-06-30 | | Release date: | 2014-07-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structures of Kibdelomycin Bound to Staphylococcus Aureus Gyrb and Pare Showed a Novel U-Shaped Binding Mode.
Acs Chem.Biol., 9, 2014
|
|
7L6G
 
 | | MbnP from Methylosinus trichosporium | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Manesis, A.C, Rosenzweig, A.C. | | Deposit date: | 2020-12-23 | | Release date: | 2021-05-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Copper binding by a unique family of metalloproteins is dependent on kynurenine formation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8H13
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x2 Disulfide (G400C and V969C), Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-10-19 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
7KZ3
 
 | | Crystal structure of KabA from Bacillus cereus UW85 in complex with the internal aldimine | | Descriptor: | 1,2-ETHANEDIOL, Aminotransferase class I/II-fold pyridoxal phosphate-dependent enzyme, SODIUM ION | | Authors: | Prasertanan, T, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2020-12-09 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Snapshots along the catalytic path of KabA, a PLP-dependent aminotransferase required for kanosamine biosynthesis in Bacillus cereus UW85.
J.Struct.Biol., 213, 2021
|
|
7KZ5
 
 | | Crystal structure of KabA from Bacillus cereus UW85 in complex with the plp external aldimine adduct with kanosamine-6-phosphate | | Descriptor: | 1,2-ETHANEDIOL, 3-deoxy-3-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-6-O-phosphono-alpha-D-gluco pyranose, Aminotransferase class I/II-fold pyridoxal phosphate-dependent enzyme, ... | | Authors: | Prasertanan, T, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2020-12-10 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Snapshots along the catalytic path of KabA, a PLP-dependent aminotransferase required for kanosamine biosynthesis in Bacillus cereus UW85.
J.Struct.Biol., 213, 2021
|
|
4W8I
 
 | | Crystal structure of LpSPL/Lpp2128, Legionella pneumophila sphingosine-1 phosphate lyase | | Descriptor: | Probable sphingosine-1-phosphate lyase | | Authors: | Stogios, P.J, Daniels, C, Skarina, T, Cuff, M, Di Leo, R, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-24 | | Release date: | 2014-11-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Legionella pneumophila S1P-lyase targets host sphingolipid metabolism and restrains autophagy.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4W9C
 
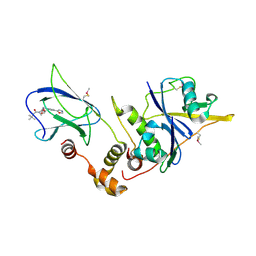 | | pVHL:EloB:EloC in complex with (2S,4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(oxazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 2) | | Descriptor: | (4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-[4-(1,3-oxazol-5-yl)benzyl]-L-prolinamide, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | Authors: | Gadd, M.S, Hewitt, S, Galdeano, C, van Molle, I, Ciulli, A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Design and Optimization of Small Molecules Targeting the Protein-Protein Interaction between the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase and the Hypoxia Inducible Factor (HIF) Alpha Subunit with in Vitro Nanomolar Affinities.
J.Med.Chem., 57, 2014
|
|
8H16
 
 | | Structure of SARS-CoV-1 Spike Protein (S/native) at pH 5.5, Open Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.35534 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H0Y
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-112 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, LINOLEIC ACID, ... | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
5LE5
 
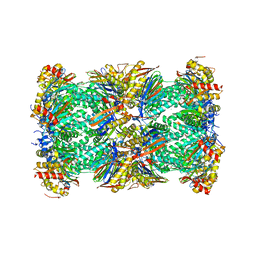 | | Native human 20S proteasome at 1.8 Angstrom | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Schrader, J, Henneberg, F, Mata, R, Tittmann, K, Schneider, T.R, Stark, H, Bourenkov, G, Chari, A. | | Deposit date: | 2016-06-29 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The inhibition mechanism of human 20S proteasomes enables next-generation inhibitor design.
Science, 353, 2016
|
|
