6OO3
 
 | | Cryo-EM structure of the C4-symmetric TRPV2/RTx complex in amphipol resolved to 2.9 A | | Descriptor: | TRPV2, resiniferatoxin | | Authors: | Zubcevic, L, Hsu, A.L, Borgnia, M.J, Lee, S.-Y. | | Deposit date: | 2019-04-22 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Symmetry transitions during gating of the TRPV2 ion channel in lipid membranes.
Elife, 8, 2019
|
|
6OPT
 
 | | HIV-1 Protease NL4-3 V82F, I84V Mutant in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Henes, M, Kosovrasti, K, Leidner, F, Nachum, G.S, Nalivaika, E.A, Bolon, D.N.A, KurtYilmaz, N, Schiffer, C.A. | | Deposit date: | 2019-04-25 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Picomolar to Micromolar: Elucidating the Role of Distal Mutations in HIV-1 Protease in Conferring Drug Resistance.
Acs Chem.Biol., 14, 2019
|
|
6OPX
 
 | | HIV-1 Protease NL4-3 I13V, G16E, V32I, L33F, K45I, M46I, L76V, V82F, I84V Mutant in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease NL4-3 | | Authors: | Lockbaum, G.J, Henes, M, Kosovrasti, K, Leidner, F, Nachum, G.S, Nalivaika, E.A, Bolon, D.N.A, KurtYilmaz, N, Schiffer, C.A. | | Deposit date: | 2019-04-25 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Picomolar to Micromolar: Elucidating the Role of Distal Mutations in HIV-1 Protease in Conferring Drug Resistance.
Acs Chem.Biol., 14, 2019
|
|
6OQ3
 
 | |
6OQE
 
 | |
6OQS
 
 | | E. coli ATP synthase State 1b | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Stewart, A.G, Sobti, M, Walshe, J.L. | | Deposit date: | 2019-04-29 | | Release date: | 2020-06-03 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures provide insight into how E. coli F1FoATP synthase accommodates symmetry mismatch.
Nat Commun, 11, 2020
|
|
6OSJ
 
 | |
6OI1
 
 | | Crystal structure of human WDR5 in complex with monomethyl L-arginine | | Descriptor: | (2S)-2-amino-5-[(N-methylcarbamimidoyl)amino]pentanoic acid, GLYCEROL, SULFATE ION, ... | | Authors: | Lorton, B.M, Harijan, R.K, Burgos, E, Bonanno, J.B, Almo, S.C, Shechter, D. | | Deposit date: | 2019-04-08 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | A Binary Arginine Methylation Switch on Histone H3 Arginine 2 Regulates Its Interaction with WDR5.
Biochemistry, 59, 2020
|
|
6OI7
 
 | | Se-Met structure of apo- Escherichia coli dGTPase | | Descriptor: | Deoxyguanosinetriphosphate triphosphohydrolase, MANGANESE (II) ION, SULFATE ION | | Authors: | Calero, G, Barnes, C.O, Wu, Y. | | Deposit date: | 2019-04-08 | | Release date: | 2019-05-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of dGTPase reveals the molecular basis of dGTP selectivity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OVI
 
 | |
3CUI
 
 | |
6OVL
 
 | |
6OWZ
 
 | | Spy H96L:Im7 L19pI-Phe complex; multiple anomalous datasets contained herein for element identification | | Descriptor: | CHLORIDE ION, IMIDAZOLE, IODIDE ION, ... | | Authors: | Rocchio, S, Duman, R, El Omari, K, Mykhaylyk, V, Yan, Z, Wagner, A, Bardwell, J.C.A, Horowitz, S. | | Deposit date: | 2019-05-12 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Identifying dynamic, partially occupied residues using anomalous scattering.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
3K1U
 
 | | Beta-xylosidase, family 43 glycosyl hydrolase from Clostridium acetobutylicum | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-xylosidase, ... | | Authors: | Osipiuk, J, Wu, R, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-28 | | Release date: | 2009-10-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray crystal structure of
beta-xylosidase, family 43 glycosyl hydrolase from Clostridium acetobutylicum at 1.55 A resolution
To be Published
|
|
6OQB
 
 | | Co-crystal structure of Mcl1 with inhibitor 10 | | Descriptor: | (4S,7aR,9aR,10S,11E,15R)-6'-chloro-15-ethyl-10-hydroxy-3',4',7a,8,9,9a,10,13,14,15-decahydro-2'H,3H,5H-spiro[1,19-(ethanediylidene)-16lambda~6~-cyclobuta[i][1,4]oxazepino[3,4-f][1,2,7]thiadiazacyclohexadecine-4,1'-naphthalene]-16,16,18(7H,17H)-trione, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Huang, X. | | Deposit date: | 2019-04-26 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | AMG 176, a Selective MCL1 Inhibitor, Is Effective in Hematologic Cancer Models Alone and in Combination with Established Therapies.
Cancer Discov, 8, 2018
|
|
3D3D
 
 | | Bacteriophage lambda lysozyme complexed with a chitohexasaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysozyme, SULFATE ION | | Authors: | Leung, A.K.W, Berghuis, A.M. | | Deposit date: | 2008-05-09 | | Release date: | 2008-09-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the lytic transglycosylase from bacteriophage lambda in complex with hexa-N-acetylchitohexaose
Biochemistry, 40, 2001
|
|
3KL3
 
 | | Crystal structure of Ligand bound XynC | | Descriptor: | D-HISTIDINE, Glucuronoxylanase xynC, TETRAETHYLENE GLYCOL, ... | | Authors: | St John, F.J, Hurlbert, J.C, Pozharski, E. | | Deposit date: | 2009-11-06 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Ligand bound structures of a glycosyl hydrolase family 30 glucuronoxylan xylanohydrolase.
J.Mol.Biol., 407, 2011
|
|
3KB6
 
 | | Crystal structure of D-Lactate dehydrogenase from aquifex aeolicus complexed with NAD and Lactic acid | | Descriptor: | D-lactate dehydrogenase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Antonyuk, S.V, Strange, R.W, Ellis, M.J, Bessho, Y, Kuramitsu, S, Yokoyama, S, Hasnain, S.S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-10-20 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure of D-lactate dehydrogenase from Aquifex aeolicus complexed with NAD(+) and lactic acid (or pyruvate).
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3KBB
 
 | | Crystal structure of putative beta-phosphoglucomutase from Thermotoga maritima | | Descriptor: | GLYCEROL, Phosphorylated carbohydrates phosphatase TM_1254, SULFATE ION | | Authors: | Strange, R.W, Antonyuk, S.V, Ellis, M.J, Bessho, Y, Kuramitsu, S, Yokoyama, S, Hasnain, S.S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-10-20 | | Release date: | 2009-11-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structure of a putative beta-phosphoglucomutase (TM1254) from Thermotoga maritima.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3CSY
 
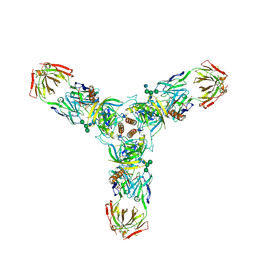 | | Crystal structure of the trimeric prefusion Ebola virus glycoprotein in complex with a neutralizing antibody from a human survivor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein GP1, ... | | Authors: | Lee, J.E, Fusco, M.L, Hessell, A.J, Oswald, W.B, Burton, D.R, Saphire, E.O. | | Deposit date: | 2008-04-10 | | Release date: | 2008-07-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor.
Nature, 454, 2008
|
|
3KLD
 
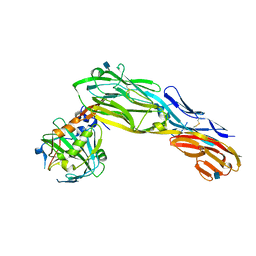 | | PTPRG CNTN4 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Contactin 4, Receptor-type tyrosine-protein phosphatase gamma | | Authors: | Bouyain, S. | | Deposit date: | 2009-11-07 | | Release date: | 2009-12-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | The protein tyrosine phosphatases PTPRZ and PTPRG bind to distinct members of the contactin family of neural recognition molecules.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3L02
 
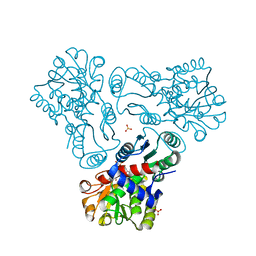 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase E92A mutant complexed with carbamyl phosphate and N-succinyl-L-norvaline | | Descriptor: | N-(3-CARBOXYPROPANOYL)-L-NORVALINE, N-acetylornithine carbamoyltransferase, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER, ... | | Authors: | Shi, D, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2009-12-09 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A single mutation in the active site swaps the substrate specificity of N-acetyl-L-ornithine transcarbamylase and N-succinyl-L-ornithine transcarbamylase.
Protein Sci., 16, 2007
|
|
3K2H
 
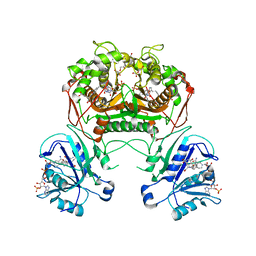 | | Co-crystal structure of dihydrofolate reductase/thymidylate synthase from Babesia bovis with dUMP, Pemetrexed and NADP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-{4-[2-(2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDIN-5-YL)-ETHYL]-BENZOYLAMINO}-PENTANEDIOIC ACID, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-09-30 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibitor-bound complexes of dihydrofolate reductase-thymidylate synthase from Babesia bovis.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3K2Z
 
 | |
3K6O
 
 | |
