6ZTO
 
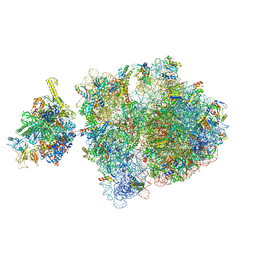 | | E. coli 70S-RNAP expressome complex in uncoupled state 1 | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
8J07
 
 | |
6YA9
 
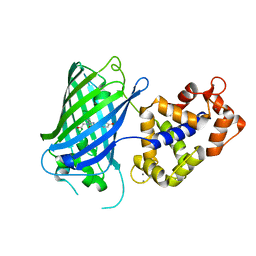 | | Crystal structure of rsGCaMP in the ON state (non-illuminated) | | Descriptor: | CALCIUM ION, rsCGaMP | | Authors: | Janowski, R, Fuenzalida-Werner, J.P, Mishra, K, Stiel, A.C, Niessing, D. | | Deposit date: | 2020-03-11 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Genetically encoded photo-switchable molecular sensors for optoacoustic and super-resolution imaging.
Nat.Biotechnol., 2021
|
|
8JFK
 
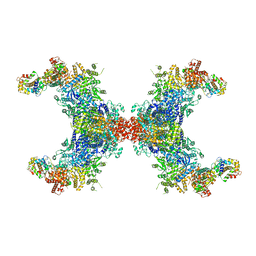 | | PhK holoenzyme in inactive state, muscle isoform | | Descriptor: | Calmodulin-1, FARNESYL, Phosphorylase b kinase gamma catalytic chain, ... | | Authors: | Yang, X.K, Xiao, J.Y. | | Deposit date: | 2023-05-18 | | Release date: | 2024-04-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Architecture and activation of human muscle phosphorylase kinase.
Nat Commun, 15, 2024
|
|
2LL6
 
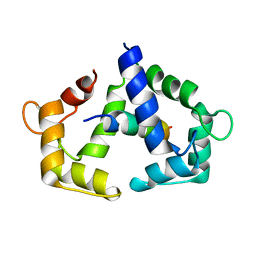 | | Solution NMR structure of CaM bound to iNOS CaM binding domain peptide | | Descriptor: | Calmodulin, Nitric oxide synthase, inducible | | Authors: | Piazza, M, Futrega, K, Spratt, D.E, Dieckmann, T, Guillemette, J.G. | | Deposit date: | 2011-10-29 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of Calmodulin (CaM) Bound to Nitric Oxide Synthase Peptides: Effects of a Phosphomimetic CaM Mutation.
Biochemistry, 51, 2012
|
|
7C8E
 
 | | Crystal Structure of 14-3-3 epsilon with 9J10 peptide | | Descriptor: | 14-3-3 protein epsilon, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 9J10 | | Authors: | Mathivanan, S, Sudhakar, S, Bairy, S, Kamariah, N, Venkitaraman, A. | | Deposit date: | 2020-05-30 | | Release date: | 2021-06-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Target identification for small-molecule discovery in the FOXO3a tumor-suppressor pathway using a biodiverse peptide library.
Cell Chem Biol, 28, 2021
|
|
5DOW
 
 | |
8OE4
 
 | | Cryo-EM structure of a pre-dimerized human IL-23 complete extracellular signaling complex. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-12 receptor subunit beta-1,Death-associated protein kinase 1, ... | | Authors: | Bloch, Y, Felix, J, Savvides, S.N. | | Deposit date: | 2023-03-10 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of complete extracellular receptor assemblies mediated by IL-12 and IL-23.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7SC0
 
 | |
7V9B
 
 | | Crystal Structure of 14-3-3 epsilon with FOXO3a peptide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[3,6-bis(dimethylamino)xanthen-9-yl]-5-methanoyl-benzoate, ARG-ARG-ARG-ALA-VAL-SEP-MET-ASP-ASN-SER-ASN, ... | | Authors: | Mathivanan, S, Kamariah, N. | | Deposit date: | 2021-08-24 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of a 14-3-3 epsilon :FOXO3a pS253 Phosphopeptide Complex Reveals 14-3-3 Isoform-Specific Binding of Forkhead Box Class O Transcription Factor (FOXO) Phosphoproteins.
Acs Omega, 7, 2022
|
|
7T2Q
 
 | | PEGylated Calmodulin-1 (K148U) | | Descriptor: | CALCIUM ION, Calmodulin-1, MAGNESIUM ION, ... | | Authors: | Mackay, J.P, Payne, R.J, Patel, K, Dowman, L.J. | | Deposit date: | 2021-12-06 | | Release date: | 2022-10-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Site-selective photocatalytic functionalization of peptides and proteins at selenocysteine.
Nat Commun, 13, 2022
|
|
5NIN
 
 | |
4X3H
 
 | | CRYSTAL STRUCTURE OF ARC N-LOBE COMPLEXED WITH STARGAZIN PEPTIDE | | Descriptor: | Activity-regulated cytoskeleton-associated protein, VOLTAGE-DEPENDENT CALCIUM CHANNEL GAMMA-2 SUBUNIT | | Authors: | zhang, W, ward, m, leahy, d, worley, p. | | Deposit date: | 2014-11-30 | | Release date: | 2015-06-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural basis of arc binding to synaptic proteins: implications for cognitive disease.
Neuron, 86, 2015
|
|
8DJH
 
 | | Ternary complex of SUMO1 with a phosphomimetic SIM of PML and zinc | | Descriptor: | PML 4SD, Small ubiquitin-related modifier 1, ZINC ION | | Authors: | Lussier-Price, M, Wahba, H.M, Mascle, X.H, Cappadocia, L, Bourdeau, V, Gagnon, C, Igelmann, S, Sakaguchi, K, Ferbeyre, G, Omichinski, J.G. | | Deposit date: | 2022-06-30 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Zinc controls PML nuclear body formation through regulation of a paralog specific auto-inhibition in SUMO1.
Nucleic Acids Res., 50, 2022
|
|
8DJI
 
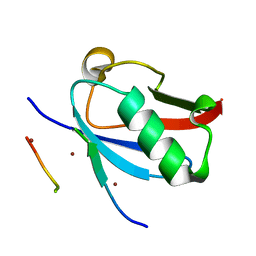 | | Ternary complex of SUMO1 with the SIM of PML and zinc | | Descriptor: | Protein PML, Small ubiquitin-related modifier 1, ZINC ION | | Authors: | Lussier-Price, M, Wahba, H.M, Mascle, X.H, Cappadocia, L, Bourdeau, V, Gagnon, C, Igelmann, S, Sakaguchi, K, Ferbeyre, G, Omichinski, J.G. | | Deposit date: | 2022-06-30 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Zinc controls PML nuclear body formation through regulation of a paralog specific auto-inhibition in SUMO1.
Nucleic Acids Res., 50, 2022
|
|
3J96
 
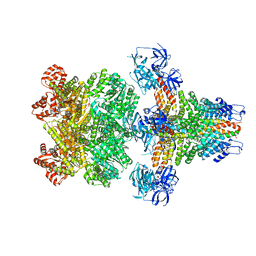 | | Structure of 20S supercomplex determined by single particle cryoelectron microscopy (State I) | | Descriptor: | Alpha-soluble NSF attachment protein, Synaptosomal-associated protein 25, Syntaxin-1A, ... | | Authors: | Zhao, M, Wu, S, Cheng, Y, Brunger, A.T. | | Deposit date: | 2014-12-05 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Mechanistic insights into the recycling machine of the SNARE complex.
Nature, 518, 2015
|
|
3J97
 
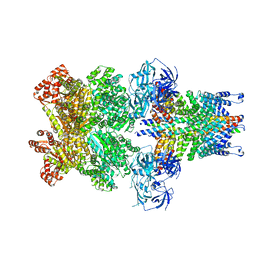 | | Structure of 20S supercomplex determined by single particle cryoelectron microscopy (State II) | | Descriptor: | Alpha-soluble NSF attachment protein, Synaptosomal-associated protein 25, Syntaxin-1A, ... | | Authors: | Zhao, M, Wu, S, Cheng, Y, Brunger, A.T. | | Deposit date: | 2014-12-05 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Mechanistic insights into the recycling machine of the SNARE complex.
Nature, 518, 2015
|
|
3J98
 
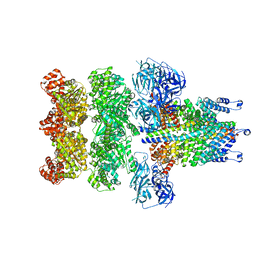 | | Structure of 20S supercomplex determined by single particle cryoelectron microscopy (State IIIa) | | Descriptor: | Alpha-soluble NSF attachment protein, Synaptosomal-associated protein 25, Syntaxin-1A, ... | | Authors: | Zhao, M, Wu, S, Cheng, Y, Brunger, A.T. | | Deposit date: | 2014-12-05 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | Mechanistic insights into the recycling machine of the SNARE complex.
Nature, 518, 2015
|
|
5MDN
 
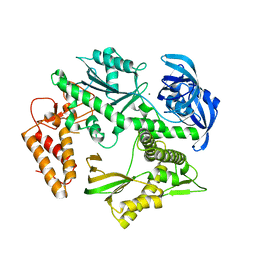 | | Structure of the family B DNA polymerase from the hyperthermophilic archaeon Pyrobaculum calidifontis | | Descriptor: | DNA polymerase, MAGNESIUM ION | | Authors: | Guo, J, Zhang, W, Coker, A.R, Wood, S.P, Cooper, J.B, Rashid, N, Akhtar, M. | | Deposit date: | 2016-11-12 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the family B DNA polymerase from the hyperthermophilic archaeon Pyrobaculum calidifontis.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
8AHS
 
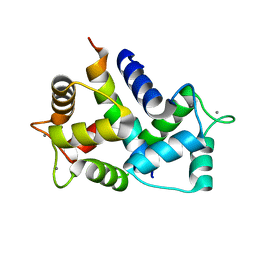 | |
3J99
 
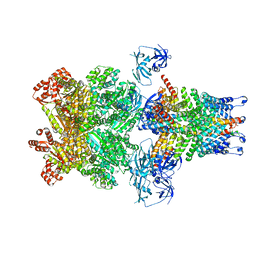 | | Structure of 20S supercomplex determined by single particle cryoelectron microscopy (State IIIb) | | Descriptor: | Alpha-soluble NSF attachment protein, Synaptosomal-associated protein 25, Syntaxin-1A, ... | | Authors: | Zhao, M, Wu, S, Cheng, Y, Brunger, A.T. | | Deposit date: | 2014-12-05 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Mechanistic insights into the recycling machine of the SNARE complex.
Nature, 518, 2015
|
|
5LDV
 
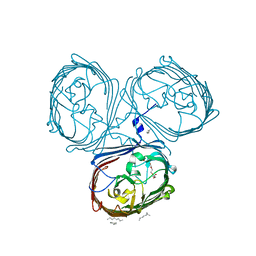 | | Crystal Structures of MOMP from Campylobacter jejuni | | Descriptor: | CALCIUM ION, MOMP porin, N-OCTANE, ... | | Authors: | Wallat, G.D, Ferrara, L.M.G, Moynie, L, Naismith, J.H. | | Deposit date: | 2016-06-28 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | MOMP from Campylobacter jejuni Is a Trimer of 18-Stranded beta-Barrel Monomers with a Ca(2+) Ion Bound at the Constriction Zone.
J.Mol.Biol., 428, 2016
|
|
7W5Z
 
 | | Cryo-EM structure of Tetrahymena thermophila mitochondrial complex IV, composite dimer model | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-oxoglutarate/malate carrier protein, CARDIOLIPIN, ... | | Authors: | Zhou, L, Maldonado, M, Padavannil, A, Letts, J. | | Deposit date: | 2021-11-30 | | Release date: | 2022-04-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structures of Tetrahymena 's respiratory chain reveal the diversity of eukaryotic core metabolism.
Science, 376, 2022
|
|
2N1V
 
 | |
2KQS
 
 | |
