5YNA
 
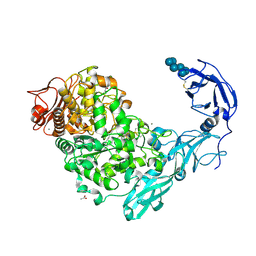 | | Crystal structure of Pullulanase from Klebsiella pneumoniae complex at 1 mM alpha-cyclodextrin | | Descriptor: | ACETATE ION, CALCIUM ION, Cyclohexakis-(1-4)-(alpha-D-glucopyranose), ... | | Authors: | Saka, N, Iwamoto, H, Takahashi, N, Mizutani, K, Mikami, B. | | Deposit date: | 2017-10-24 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Elucidation of the mechanism of interaction between Klebsiella pneumoniae pullulanase and cyclodextrin
Acta Crystallogr D Struct Biol, 74, 2018
|
|
3TAO
 
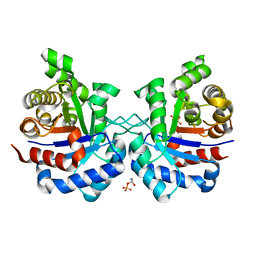 | | Structure of Mycobacterium tuberculosis triosephosphate isomerase bound to phosphoglycolohydroxamate | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, Triosephosphate isomerase | | Authors: | Connor, S.E, Capodagli, G.C, Deaton, M.K, Pegan, S.D. | | Deposit date: | 2011-08-04 | | Release date: | 2011-11-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and functional characterization of Mycobacterium tuberculosis triosephosphate isomerase.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3TA6
 
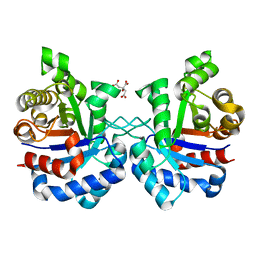 | | Structure of Mycobacterium tuberculosis triosephosphate isomerase | | Descriptor: | CITRATE ANION, Triosephosphate isomerase | | Authors: | Connor, S.E, Capodagli, G.C, Deaton, M.K, Pegan, S.D. | | Deposit date: | 2011-08-03 | | Release date: | 2011-11-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural and functional characterization of Mycobacterium tuberculosis triosephosphate isomerase.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
4GWT
 
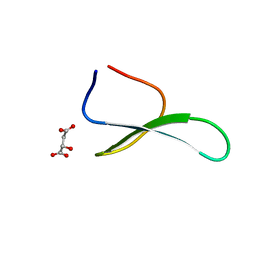 | | Structure of racemic Pin1 WW domain cocrystallized with DL-malic acid | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Mortenson, D.E, Yun, H.G, Gellman, S.H, Forest, K.T. | | Deposit date: | 2012-09-03 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Evidence for small-molecule-mediated loop stabilization in the structure of the isolated Pin1 WW domain.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5YIF
 
 | | Pyruvylated beta-D-galactosidase from Bacillus sp. HMA207, E163A mutant pyruvylated beta-D-galactose complex | | Descriptor: | (2R,4aR,6R,7R,8R,8aR)-2-methyl-6,7,8-tris(oxidanyl)-4,4a,6,7,8,8a-hexahydropyrano[3,2-d][1,3]dioxine-2-carboxylic acid, Pyruvylated beta-D-galactosidase | | Authors: | Tanuma, M, Yamada, C, Arakawa, T, Higuchi, Y, Takegawa, K, Fushinobu, S. | | Deposit date: | 2017-10-04 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Identification and characterization of a novel beta-D-galactosidase that releases pyruvylated galactose.
Sci Rep, 8, 2018
|
|
5YNC
 
 | | Crystal structure of Pullulanase from Klebsiella pneumoniae complex at 1 mM beta-cyclodextrin | | Descriptor: | ACETATE ION, CALCIUM ION, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), ... | | Authors: | Saka, N, Iwamoto, H, Takahashi, N, Mizutani, K, Mikami, B. | | Deposit date: | 2017-10-24 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Elucidation of the mechanism of interaction between Klebsiella pneumoniae pullulanase and cyclodextrin
Acta Crystallogr D Struct Biol, 74, 2018
|
|
4III
 
 | | Lambda-[Ru(TAP)2(11-Cl-dppz)] with a DNA decamer at atomic resolution | | Descriptor: | 5'-D(*TP*CP*GP*GP*CP*GP*CP*CP*GP*A)-3', BARIUM ION, Ru(TAP)2(Cl-dppz) complex | | Authors: | Hall, J.P, Cardin, C.J. | | Deposit date: | 2012-12-20 | | Release date: | 2013-07-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Preferred orientation in an angled intercalation site of a chloro-substituted Lambda-[Ru(TAP)2(dppz)]2+ complex bound to d(TCGGCGCCGA)2.
Philos Trans A Math Phys Eng Sci, 371, 2013
|
|
5G2V
 
 | | Structure of BT4656 in complex with its substrate D-Glucosamine-2-N, 6-O-disulfate. | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CALCIUM ION, N-ACETYLGLUCOSAMINE-6-SULFATASE, ... | | Authors: | Cartmell, A, Lowe, E.C, Basle, A, Crouch, L.I, Czjzek, M, Turnbull, J, Henrissat, B, Terrapon, N, Thomas, S, Murray, H, Firbank, S.J, Bolam, D.N. | | Deposit date: | 2016-04-14 | | Release date: | 2017-05-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | How members of the human gut microbiota overcome the sulfation problem posed by glycosaminoglycans.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3RZT
 
 | | Neutron structure of perdeuterated rubredoxin using rapid (14 hours) data | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Munshi, P, Chung, C.-L, Weiss, K.L, Blakeley, M.P, Myles, D.A.A, Meilleur, F. | | Deposit date: | 2011-05-12 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | NEUTRON DIFFRACTION (1.7504 Å) | | Cite: | Rapid visualization of hydrogen positions in protein neutron crystallographic structures.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3RZI
 
 | | The structure of 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase from mycobacterium tuberculosis cocrystallized and complexed with phenylalanine and tryptophan | | Descriptor: | CHLORIDE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Jiao, W, Jameson, G.B, Hutton, R.D, Parker, E.J. | | Deposit date: | 2011-05-11 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Dynamic cross-talk among remote binding sites: the molecular basis for unusual synergistic allostery.
J.Mol.Biol., 415, 2012
|
|
2WYN
 
 | | Structure of family 37 trehalase from Escherichia coli in complex with a casuarine-6-O-a-D-glucoside analogue | | Descriptor: | (1R,2R,3R,6R,7R,7AR)-3,7-BIS(HYDROXYMETHYL)HEXAHYDRO-1H-PYRROLIZINE-1,2,6-TRIOL, CALCIUM ION, PERIPLASMIC TREHALASE, ... | | Authors: | Gloster, T.M, Roberts, S.M, Davies, G.J, Cardona, F, Goti, A, Parmeggiani, C, Parenti, P, Fusi, P, Forcella, M, Cipolla, L. | | Deposit date: | 2009-11-17 | | Release date: | 2010-09-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Casuarine-6-O-alpha-D-glucoside and its analogues are tight binding inhibitors of insect and bacterial trehalases.
Chem.Commun.(Camb.), 46, 2010
|
|
5YNE
 
 | | Crystal structure of Pullulanase from Klebsiella pneumoniae complex at 10 mM alpha-cyclodextrin | | Descriptor: | CALCIUM ION, Cyclohexakis-(1-4)-(alpha-D-glucopyranose), DI(HYDROXYETHYL)ETHER, ... | | Authors: | Saka, N, Iwamoto, H, Takahashi, N, Mizutani, K, Mikami, B. | | Deposit date: | 2017-10-24 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Elucidation of the mechanism of interaction between Klebsiella pneumoniae pullulanase and cyclodextrin
Acta Crystallogr D Struct Biol, 74, 2018
|
|
3P0W
 
 | | Crystal structure of D-Glucarate dehydratase from Ralstonia solanacearum complexed with Mg and D-glucarate | | Descriptor: | D-GLUCARATE, MAGNESIUM ION, Mandelate racemase/muconate lactonizing protein | | Authors: | Fedorov, A.A, Fedorov, E.V, Gerlt, J.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-09-29 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal structure of D-Glucarate dehydratase from Ralstonia solanacearum complexed with Mg and D-glucarate
To be Published
|
|
3B8X
 
 | | Crystal structure of GDP-4-keto-6-deoxymannose-3-dehydratase (ColD) H188N mutant with bound GDP-perosamine | | Descriptor: | 1,2-ETHANEDIOL, Pyridoxamine 5-phosphate-dependent dehydrase, SODIUM ION, ... | | Authors: | Cook, P.D, Holden, H.M. | | Deposit date: | 2007-11-02 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | GDP-4-Keto-6-deoxy-D-mannose 3-Dehydratase, Accommodating a Sugar Substrate in the Active Site.
J.Biol.Chem., 283, 2008
|
|
1GQ1
 
 | | CYTOCHROME CD1 NITRITE REDUCTASE, Y25S mutant, OXIDISED FORM | | Descriptor: | CYTOCHROME CD1 NITRITE REDUCTASE, GLYCEROL, HEME C, ... | | Authors: | Sjogren, T, Gordon, E.H.J, Lofqvist, M, Richter, C.D, Hajdu, J, Ferguson, S.J. | | Deposit date: | 2001-11-19 | | Release date: | 2002-11-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and Kinetic Properties of Paracoccus Pantotrophus Cytochrome Cd1 Nitrite Reductase with the D1 Heme Active Site Ligand Tyrosine 25 Replaced by Serine
J.Biol.Chem., 278, 2003
|
|
5YN7
 
 | | Crystal structure of Pullulanase from Klebsiella pneumoniae complex at 0.1 mM beta-cyclodextrin | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), ... | | Authors: | Saka, N, Iwamoto, H, Takahashi, N, Mizutani, K, Mikami, B. | | Deposit date: | 2017-10-24 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.586 Å) | | Cite: | Elucidation of the mechanism of interaction between Klebsiella pneumoniae pullulanase and cyclodextrin
Acta Crystallogr D Struct Biol, 74, 2018
|
|
2CBJ
 
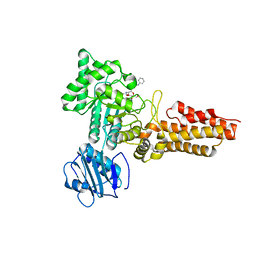 | | Structure of the Clostridium perfringens NagJ family 84 glycoside hydrolase, a homologue of human O-GlcNAcase in complex with PUGNAc | | Descriptor: | CHLORIDE ION, HYALURONIDASE, O-(2-ACETAMIDO-2-DEOXY D-GLUCOPYRANOSYLIDENE) AMINO-N-PHENYLCARBAMATE | | Authors: | Rao, F.V, Dorfmueller, H.C, Villa, F, Allwood, M, Eggleston, I.M, van Aalten, D.M.F. | | Deposit date: | 2006-01-05 | | Release date: | 2006-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insights into the mechanism and inhibition of eukaryotic O-GlcNAc hydrolysis.
EMBO J., 25, 2006
|
|
5KAF
 
 | | RT XFEL structure of Photosystem II in the dark state at 3.0 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Fuller, F, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Waterman, D.G, Evans, G, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-06-01 | | Release date: | 2016-11-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.00001 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
4RYA
 
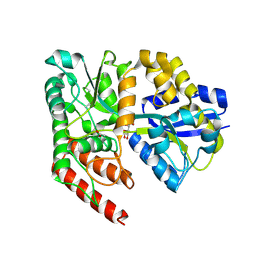 | | Crystal structure of abc transporter solute binding protein AVI_3567 from AGROBACTERIUM VITIS S4, TARGET EFI-510645, with bound D-mannitol | | Descriptor: | ABC transporter substrate binding protein (Sorbitol), ACETATE ION, D-MANNITOL | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siedel, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-13 | | Release date: | 2014-12-24 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Periplasmic Solute-Binding Protein Avi_3567 from Agrobacterium Vitis, Target Efi-510645
To be Published
|
|
4TTH
 
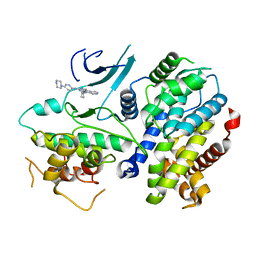 | | Crystal structure of a CDK6/Vcyclin complex with inhibitor bound | | Descriptor: | 9-cyclopentyl-N-(5-piperazin-1-ylpyridin-2-yl)pyrido[4,5]pyrrolo[1,2-d]pyrimidin-2-amine, Cyclin homolog, Cyclin-dependent kinase 6 | | Authors: | Piper, D.E, Walker, N, Wang, Z. | | Deposit date: | 2014-06-20 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of AMG 925, a FLT3 and CDK4 dual kinase inhibitor with preferential affinity for the activated state of FLT3.
J.Med.Chem., 57, 2014
|
|
1JUC
 
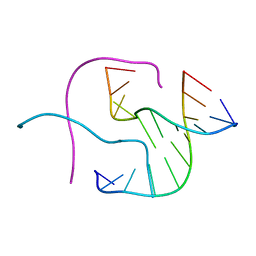 | | Crystal Structure Analysis of a Holliday Junction Formed by CCGGTACCGG | | Descriptor: | 5'-D(*CP*CP*GP*GP*TP*AP*CP*CP*GP*G)-3' | | Authors: | Thorpe, J.H, Teixeira, S.C.M, Gale, B.C, Cardin, C.J. | | Deposit date: | 2001-08-24 | | Release date: | 2002-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural characterization of a new crystal form of the four-way Holliday junction formed by the DNA sequence d(CCGGTACCGG)2: sequence versus lattice?
Acta Crystallogr.,Sect.D, 58, 2002
|
|
5KAI
 
 | | NH3-bound RT XFEL structure of Photosystem II 500 ms after the 2nd illumination (2F) at 2.8 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Fuller, F, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Waterman, D.G, Evans, G, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-06-01 | | Release date: | 2016-11-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.80000925 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
5DZU
 
 | | Structure of potato cathepsin D inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Aspartic protease inhibitor 11, ... | | Authors: | Guo, J, Erskine, P, Coker, A.R, Wood, S.P, Cooper, J.B. | | Deposit date: | 2015-09-26 | | Release date: | 2015-10-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure of a Kunitz-type potato cathepsin D inhibitor.
J.Struct.Biol., 192, 2015
|
|
5YNH
 
 | | Crystal structure of Pullulanase from Klebsiella pneumoniae complex at 10 mM gamma-cyclodextrin | | Descriptor: | CALCIUM ION, Cyclooctakis-(1-4)-(alpha-D-glucopyranose), GLYCEROL, ... | | Authors: | Saka, N, Iwamoto, H, Takahashi, N, Mizutani, K, Mikami, B. | | Deposit date: | 2017-10-24 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Elucidation of the mechanism of interaction between Klebsiella pneumoniae pullulanase and cyclodextrin
Acta Crystallogr D Struct Biol, 74, 2018
|
|
2FG4
 
 | | Structure of Human Ferritin L Chain | | Descriptor: | CADMIUM ION, Ferritin light chain | | Authors: | Wang, Z, Li, C, Ellenburg, M, Ruble, J, Ho, J.X, Carter, D.C. | | Deposit date: | 2005-12-21 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of human ferritin L chain.
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
