2FU2
 
 | | Crystal structure of protein SPy2152 from Streptococcus pyogenes | | Descriptor: | Hypothetical protein SPy2152 | | Authors: | Chang, C, Cymborowski, M, Otwinowski, Z, Minor, W, Lezondra, L.-E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-01-25 | | Release date: | 2006-03-07 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure of pyogenecin immunity protein, a novel bacteriocin-like immunity protein from Streptococcus pyogenes.
Bmc Struct.Biol., 9, 2009
|
|
5VXB
 
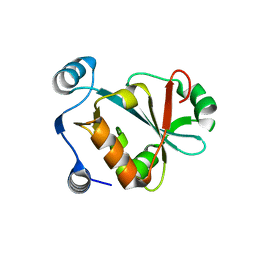 | |
4O72
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with NU7441 | | Descriptor: | 1,2-ETHANEDIOL, 8-(dibenzo[b,d]thiophen-4-yl)-2-(morpholin-4-yl)-4H-chromen-4-one, Bromodomain-containing protein 4, ... | | Authors: | Zhu, J.-Y, Ember, S.W, Watts, C, Schonbrunn, E. | | Deposit date: | 2013-12-24 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
5VNJ
 
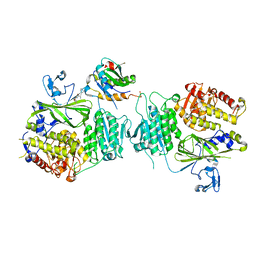 | |
2G0A
 
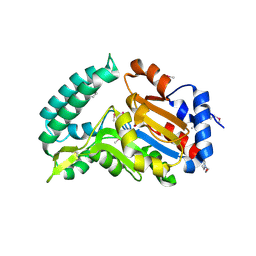 | | X-ray structure of mouse pyrimidine 5'-nucleotidase type 1 with lead(II) bound in active site | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Cytosolic 5'-nucleotidase III, LEAD (II) ION | | Authors: | Bitto, E, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-02-11 | | Release date: | 2006-04-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of pyrimidine 5'-nucleotidase type 1. Insight into mechanism of action and inhibition during lead poisoning.
J.Biol.Chem., 281, 2006
|
|
5VOK
 
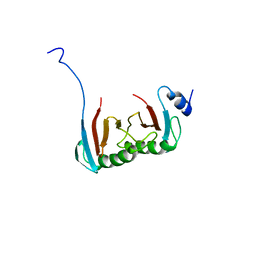 | | Crystal structure of the C7orf59-HBXIP dimer | | Descriptor: | Ragulator complex protein LAMTOR4, Ragulator complex protein LAMTOR5 | | Authors: | Rasheed, N, Nascimento, A.F.Z, Bar-Peled, L, Shen, K, Sabatini, D.M, Aparicio, R, Smetana, J.H.C. | | Deposit date: | 2017-05-03 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | C7orf59/Lamtor4 phosphorylation and structural flexibility modulate Ragulator assembly.
Febs Open Bio, 2019
|
|
4O70
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with DINACICLIB | | Descriptor: | 1,2-ETHANEDIOL, 3-[({3-ethyl-5-[(2S)-2-(2-hydroxyethyl)piperidin-1-yl]pyrazolo[1,5-a]pyrimidin-7-yl}amino)methyl]-1-hydroxypyridinium, Bromodomain-containing protein 4 | | Authors: | Ember, S.W, Zhu, J.-Y, Watts, C, Schonbrunn, E. | | Deposit date: | 2013-12-24 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
8P98
 
 | | BtuB3G3 bound to cyanocobalamin with ordered EL8 | | Descriptor: | CYANOCOBALAMIN, Putative surface layer protein, Vitamin B12 transporter BtuB | | Authors: | Silale, A, Abellon-Ruiz, J, van den Berg, B. | | Deposit date: | 2023-06-05 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | BtuB TonB-dependent transporters and BtuG surface lipoproteins form stable complexes for vitamin B 12 uptake in gut Bacteroides.
Nat Commun, 14, 2023
|
|
4O7F
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with SB-251527 | | Descriptor: | 1,2-ETHANEDIOL, 4-[4-(4-fluorophenyl)-1-(piperidin-4-yl)-1H-imidazol-5-yl]-2-(2-methoxyphenoxy)pyrimidine, Bromodomain-containing protein 4, ... | | Authors: | Ember, S.W, Zhu, J.-Y, Watts, C, Schonbrunn, E. | | Deposit date: | 2013-12-24 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
5VOY
 
 | | Yeast V-ATPase in complex with Legionella pneumophila effector SidK (rotational state 2) | | Descriptor: | V-type proton ATPase catalytic subunit A,V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Zhao, J. | | Deposit date: | 2017-05-03 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Molecular basis for the binding and modulation of V-ATPase by a bacterial effector protein.
PLoS Pathog., 13, 2017
|
|
3KR2
 
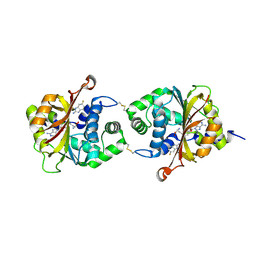 | |
3KT0
 
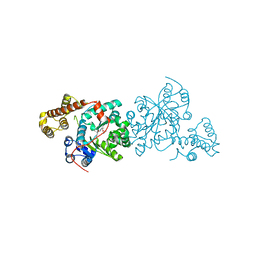 | | Crystal structure of S. cerevisiae tryptophanyl-tRNA synthetase | | Descriptor: | (2S)-1-{[(2S)-3-(2-methoxyethoxy)-2-methylpropyl]oxy}propan-2-amine, Tryptophanyl-tRNA synthetase, cytoplasmic | | Authors: | Zhou, M, Dong, X, Zhong, C, Shen, N, Ding, J. | | Deposit date: | 2009-11-24 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of Saccharomyces cerevisiae tryptophanyl-tRNA synthetase: new insights into the mechanism of tryptophan activation and implications for anti-fungal drug design
Nucleic Acids Res., 38, 2010
|
|
5VST
 
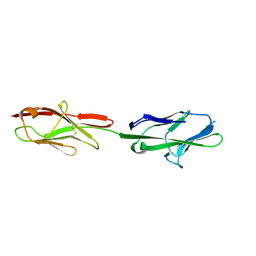 | | Crystal structure of murine CEACAM1b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Biliary glycoprotein | | Authors: | Peng, G, Yang, Y, Pasquarella, J.R, Xu, L, Qian, Z, Holmes, K.V, Li, F. | | Deposit date: | 2017-05-12 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular mechanism for coronavirus-driven evolution of mouse receptor
J. Biol. Chem., 292, 2017
|
|
5VU2
 
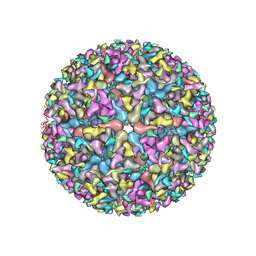 | |
4OG8
 
 | | Human menin with bound inhibitor MIV-6R | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, 4-(3-{4-[(R)-amino(cyclopentyl)phenylmethyl]piperidin-1-yl}propoxy)benzonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | He, S, Senter, T.J, Pollock, J.W, Han, C, Upadhyay, S.K, Purohit, T, Gogliotti, R.D, Lindsley, C.W, Cierpicki, T, Stauffer, S.R, Grembecka, J. | | Deposit date: | 2014-01-15 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | High-Affinity Small-Molecule Inhibitors of the Menin-Mixed Lineage Leukemia (MLL) Interaction Closely Mimic a Natural Protein-Protein Interaction.
J.Med.Chem., 57, 2014
|
|
2G9J
 
 | | Complex of TM1a(1-14)Zip with TM9a(251-284): a model for the polymerization domain ("overlap region") of tropomyosin, Northeast Structural Genomics Target OR9 | | Descriptor: | Tropomyosin 1 alpha chain, Tropomyosin 1 alpha chain/General control protein GCN4 | | Authors: | Greenfield, N.J, Huang, Y.J, Swapna, G.V.T, Bhattacharya, A, Singh, A, Montelione, G.T, Hitchcock-DeGregori, S.E, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-03-06 | | Release date: | 2006-11-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the Junction between Tropomyosin Molecules: Implications for Actin Binding and Regulation.
J.Mol.Biol., 364, 2006
|
|
5W6E
 
 | | PDE1b complexed with compound 3S | | Descriptor: | 7,8-dimethoxy-N-[(2S)-1-(3-methyl-1H-pyrazol-5-yl)propan-2-yl]quinazolin-4-amine, Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B, MAGNESIUM ION, ... | | Authors: | Vajdos, F.F. | | Deposit date: | 2017-06-16 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Potent and Selective Periphery-Restricted Quinazoline Inhibitors of the Cyclic Nucleotide Phosphodiesterase PDE1.
J. Med. Chem., 61, 2018
|
|
3KZ3
 
 | |
3L3Z
 
 | | Crystal structure of DHT-bound androgen receptor in complex with the third motif of steroid receptor coactivator 3 | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, Nuclear receptor coactivator 3 | | Authors: | Zhou, X.E, Suino-Powell, K.M, Li, J, He, A, MacKeigan, J.P, Melcher, K, Yong, E.-L, Xu, H.E. | | Deposit date: | 2009-12-18 | | Release date: | 2010-01-12 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of SRC3/AIB1 as a Preferred Coactivator for Hormone-activated Androgen Receptor.
J.Biol.Chem., 285, 2010
|
|
7SJN
 
 | | HtrA1:Fab15H6.v4 complex | | Descriptor: | Fab15H6.v4 Heavy Chain, Fab15H6.v4 Light Chain, Serine protease HTRA1 | | Authors: | Gerhardy, S, Green, E, Estevez, A, Arthur, C.P, Ultsch, M, Rohou, A, Kirchhofer, D. | | Deposit date: | 2021-10-18 | | Release date: | 2022-09-07 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Allosteric inhibition of HTRA1 activity by a conformational lock mechanism to treat age-related macular degeneration.
Nat Commun, 13, 2022
|
|
5W5A
 
 | | Crystal structure of Mycobacterium tuberculosis CRP-FNR family transcription factor Cmr (Rv1675c) | | Descriptor: | CHLORIDE ION, HTH-type transcriptional regulator Cmr, SULFATE ION | | Authors: | Cheung, J, Cassidy, M, Ginter, C, Ranganathan, S, Pata, D.J, McDonough, K.A. | | Deposit date: | 2017-06-14 | | Release date: | 2017-12-13 | | Last modified: | 2019-01-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Novel structural features drive DNA binding properties of Cmr, a CRP family protein in TB complex mycobacteria.
Nucleic Acids Res., 46, 2018
|
|
2FWU
 
 | |
4ONF
 
 | | Fab fragment of 3D6 in complex with amyloid beta 1-7 | | Descriptor: | 3D6 FAB ANTIBODY HEAVY CHAIN, 3D6 FAB ANTIBODY LIGHT CHAIN, Amyloid beta A4 protein | | Authors: | Feinberg, H, Saldanha, J.W, Diep, L, Goel, A, Widom, A, Veldman, G.M, Weis, W.I, Schenk, D, Basi, G.S. | | Deposit date: | 2014-01-28 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure reveals conservation of amyloid-beta conformation recognized by 3D6 following humanization to bapineuzumab.
Alzheimers Res Ther, 6, 2014
|
|
5VT9
 
 | |
3KM3
 
 | |
