1Y5B
 
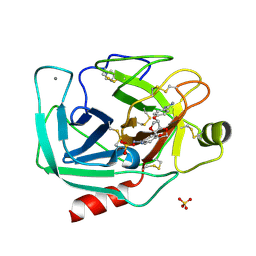 | | Dianhydrosugar-based benzamidine, factor Xa specific inhibitor in complex with bovine trypsin mutant | | Descriptor: | 2,5-BIS-O-{4-[AMINO(IMINO)METHYL]PHENYL}-1,4:3,6-DIANHYDRO-D-GLUCITOL, CALCIUM ION, IMIDAZOLE, ... | | Authors: | Di Fenza, A, Heine, A, Klebe, G. | | Deposit date: | 2004-12-02 | | Release date: | 2005-12-13 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Understanding binding selectivity toward trypsin and factor Xa: the role of aromatic interactions
Chemmedchem, 2, 2007
|
|
1YI5
 
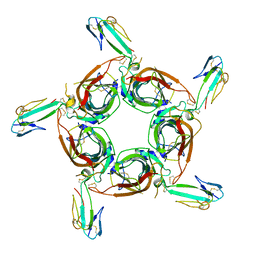 | | Crystal structure of the a-cobratoxin-AChBP complex | | Descriptor: | Acetylcholine-binding protein, Long neurotoxin 1 | | Authors: | Bourne, Y, Talley, T.T, Hansen, S.B, Taylor, P, Marchot, P. | | Deposit date: | 2005-01-11 | | Release date: | 2005-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Crystal structure of a Cbtx-AChBP complex reveals essential interactions between snake alpha-neurotoxins and nicotinic receptors
Embo J., 24, 2005
|
|
5EQ4
 
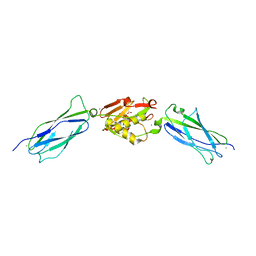 | | Crystal structure of the SrpA adhesin R347E mutant from Streptococcus sanguinis | | Descriptor: | ACETATE ION, CALCIUM ION, Platelet-binding glycoprotein | | Authors: | Loukachevitch, L.V, McCulloch, K.M, Vann, K.R, Wawrzak, Z, Anderson, S, Iverson, T.M. | | Deposit date: | 2015-11-12 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Sialoglycan Binding by the Streptococcus sanguinis SrpA Adhesin.
J.Biol.Chem., 291, 2016
|
|
5EQ2
 
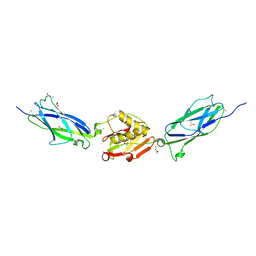 | | Crystal Structure of the SrpA Adhesin from Streptococcus sanguinis | | Descriptor: | ACETATE ION, CALCIUM ION, Platelet-binding glycoprotein | | Authors: | Loukachevitch, L.V, McCulloch, K.M, Vann, K.R, Wawrzak, Z, Anderson, S, Iverson, T.M. | | Deposit date: | 2015-11-12 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Sialoglycan Binding by the Streptococcus sanguinis SrpA Adhesin.
J.Biol.Chem., 291, 2016
|
|
5EQ3
 
 | | Crystal structure of the SrpA adhesin from Streptococcus sanguinis with a sialyl galactose disaccharide bound | | Descriptor: | ACETATE ION, CALCIUM ION, N-glycolyl-alpha-neuraminic acid-(2-3)-methyl beta-D-galactopyranoside, ... | | Authors: | Loukachevitch, L.V, McCulloch, K.M, Vann, K.R, Wawrzak, Z, Anderson, S, Iverson, T.M. | | Deposit date: | 2015-11-12 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Sialoglycan Binding by the Streptococcus sanguinis SrpA Adhesin.
J.Biol.Chem., 291, 2016
|
|
5EPL
 
 | | Crystal Structure of chromodomain of CBX4 in complex with inhibitor UNC3866 | | Descriptor: | E3 SUMO-protein ligase CBX4, UNKNOWN ATOM OR ION, unc3866 | | Authors: | Liu, Y, Tempel, W, Walker, J.R, Stuckey, J.I, Dickson, B.M, James, L.I, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-11 | | Release date: | 2015-12-23 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | A cellular chemical probe targeting the chromodomains of Polycomb repressive complex 1.
Nat.Chem.Biol., 12, 2016
|
|
7SUV
 
 | | APE1 exonuclease substrate complex with 8oxoG opposite A | | Descriptor: | DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(8OG))-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*AP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(P*TP*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), ... | | Authors: | Whitaker, A.W, Freudenthal, B.D. | | Deposit date: | 2021-11-18 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Processing oxidatively damaged bases at DNA strand breaks by APE1.
Nucleic Acids Res., 50, 2022
|
|
7SVB
 
 | | APE1 exonuclease substrate complex with 8oxoG opposite C | | Descriptor: | DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(8OG))-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*AP*CP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(P*TP*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), ... | | Authors: | Whitaker, A.W, Freudenthal, B.D. | | Deposit date: | 2021-11-18 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Processing oxidatively damaged bases at DNA strand breaks by APE1.
Nucleic Acids Res., 50, 2022
|
|
3EQ6
 
 | | Crystal structure of human acyl-CoA synthetase medium-chain family member 2A (L64P mutation) in a ternary complex with products | | Descriptor: | ADENOSINE MONOPHOSPHATE, Acyl-coenzyme A synthetase ACSM2A, Butyryl Coenzyme A | | Authors: | Pilka, E.S, Kochan, G, Yue, W.W, Bhatia, C, Von delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-09-30 | | Release date: | 2008-10-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural snapshots for the conformation-dependent catalysis by human medium-chain acyl-coenzyme A synthetase ACSM2A
J.Mol.Biol., 388, 2009
|
|
3N21
 
 | | Crystal structure of Thermolysin in complex with S-1,2-Propandiol | | Descriptor: | CALCIUM ION, S-1,2-PROPANEDIOL, Thermolysin, ... | | Authors: | Behnen, J, Heine, A, Klebe, G. | | Deposit date: | 2010-05-17 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Experimental and computational active site mapping as a starting point to fragment-based lead discovery.
Chemmedchem, 7, 2012
|
|
3EO9
 
 | | Crystal structure the Fab fragment of Efalizumab | | Descriptor: | Efalizumab Fab fragment, heavy chain, light chain | | Authors: | Li, S, Ding, J. | | Deposit date: | 2008-09-26 | | Release date: | 2009-04-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Efalizumab binding to the LFA-1 alphaL I domain blocks ICAM-1 binding via steric hindrance.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1Y6P
 
 | | Crystal structure of disulfide engineered porcine pancratic phospholipase a2 to group-x isozyme | | Descriptor: | CALCIUM ION, CHLORIDE ION, Phospholipase A2, ... | | Authors: | Yu, B.Z, Pan, Y.H, Jassen, M.J.W, Bahnson, B.J, Jain, M.K. | | Deposit date: | 2004-12-06 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Kinetic and structural properties of disulfide engineered phospholipase a(2): insight into the role of disulfide bonding patterns.
Biochemistry, 44, 2005
|
|
1K4M
 
 | | Crystal structure of E.coli nicotinic acid mononucleotide adenylyltransferase complexed to deamido-NAD | | Descriptor: | CITRIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, NaMN adenylyltransferase | | Authors: | Zhang, H, Zhou, T, Kurnasov, O, Cheek, S, Grishin, N.V, Osterman, A. | | Deposit date: | 2001-10-08 | | Release date: | 2002-10-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of E. coli nicotinate mononucleotide adenylyltransferase and its complex with deamido-NAD.
Structure, 10, 2002
|
|
2VZE
 
 | | Crystal structure of human acyl-CoA synthetase medium-chain family member 2A (L64P mutation) in complex with AMP | | Descriptor: | ACYL-COENZYME A SYNTHETASE ACSM2A, MITOCHONDRIAL, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Yue, W.W, Kochan, G.T, Pilka, E.S, Bhatia, C, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Wikstrom, M, Bountra, C, Oppermann, U. | | Deposit date: | 2008-07-31 | | Release date: | 2008-10-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Snapshots for the Conformation- Dependent Catalysis by Human Medium-Chain Acyl- Coenzyme a Synthetase Acsm2A.
J.Mol.Biol., 388, 2009
|
|
1K4K
 
 | | Crystal structure of E. coli Nicotinic acid mononucleotide adenylyltransferase | | Descriptor: | Nicotinic acid mononucleotide adenylyltransferase, XENON | | Authors: | Zhang, H, Zhou, T, Kurnasov, O, Cheek, S, Grishin, N.V, Osterman, A.L. | | Deposit date: | 2001-10-08 | | Release date: | 2002-10-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of E. coli nicotinate mononucleotide adenylyltransferase and its complex with deamido-NAD.
Structure, 10, 2002
|
|
2W1U
 
 | | A family 32 carbohydrate-binding module, from the Mu toxin produced by Clostridium perfringens, in complex with beta-D-glcNAc-beta(1,3) galNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose, ACETATE ION, ... | | Authors: | Ficko-Blean, E, Boraston, A.B. | | Deposit date: | 2008-10-20 | | Release date: | 2009-05-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | N-Acetylglucosamine Recognition by a Family 32 Carbohydrate-Binding Module from Clostridium Perfringens Nagh.
J.Mol.Biol., 390, 2009
|
|
2WDB
 
 | | A family 32 carbohydrate-binding module, from the Mu toxin produced by Clostridium perfringens, in complex with beta-D-glcNAc-beta(1,2) mannose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose, CALCIUM ION, HYALURONOGLUCOSAMINIDASE | | Authors: | Ficko-Blean, E, Boraston, A.B. | | Deposit date: | 2009-03-23 | | Release date: | 2009-05-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | N-Acetylglucosamine Recognition by a Family 32 Carbohydrate-Binding Module from Clostridium Perfringens Nagh.
J.Mol.Biol., 390, 2009
|
|
2W8Y
 
 | | RU486 bound to the progesterone receptor in a destabilized agonistic conformation | | Descriptor: | (14beta,17alpha)-17-ethynyl-17-hydroxyestr-4-en-3-one, 1,2-ETHANEDIOL, 11-(4-DIMETHYLAMINO-PHENYL)-17-HYDROXY-13-METHYL-17-PROP-1-YNYL-1,2,6,7,8,11,12,13,14,15,16,17-DODEC AHYDRO-CYCLOPENTA[A]PHENANTHREN-3-ONE, ... | | Authors: | Raaijmakers, H.C.A, Versteeg, J, Uitdehaag, J.C.M. | | Deposit date: | 2009-01-20 | | Release date: | 2009-04-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The X-Ray Structure of Ru486 Bound to the Progesterone Receptor in a Destabilized Agonistic Conformation.
J.Biol.Chem., 284, 2009
|
|
2WD9
 
 | | CRYSTAL STRUCTURE OF HUMAN ACYL-COA SYNTHETASE MEDIUM-CHAIN FAMILY MEMBER 2A (L64P MUTATION) IN COMPLEX WITH IBUPROFEN | | Descriptor: | ACYL-COENZYME A SYNTHETASE ACSM2A, MITOCHONDRIAL, IBUPROFEN, ... | | Authors: | Yue, W.W, Kochan, G.T, Pilka, E.S, Bhatia, C, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Oppermann, U. | | Deposit date: | 2009-03-21 | | Release date: | 2009-04-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Snapshots for the Conformation- Dependent Catalysis by Human Medium-Chain Acyl- Coenzyme a Synthetase Acsm2A.
J.Mol.Biol., 388, 2009
|
|
2VSS
 
 | | Wild-type Hydroxycinnamoyl-CoA hydratase lyase in complex with acetyl- CoA and vanillin | | Descriptor: | 4-hydroxy-3-methoxybenzaldehyde, ACETYL COENZYME *A, P-HYDROXYCINNAMOYL COA HYDRATASE/LYASE | | Authors: | Bennett, J.P, Bertin, L.M, Brzozowski, A.M, Walton, N.J, Grogan, G. | | Deposit date: | 2008-04-29 | | Release date: | 2008-05-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | A Ternary Complex of Hydroxycinnamoyl-Coa Hydratase-Lyase (Hchl) with Acetyl-Coa and Vanillin Gives Insights Into Substrate Specificity and Mechanism.
Biochem.J., 414, 2008
|
|
1KMY
 
 | | Crystal Structure of 2,3-dihydroxybiphenyl 1,2-dioxygenase Complexed with 2,3-dihydroxybiphenyl under Anaerobic Condition | | Descriptor: | 2,3-DIHYDROXYBIPHENYL 1,2-DIOXYGENASE, BIPHENYL-2,3-DIOL, FE (II) ION, ... | | Authors: | Han, S, Bolin, J.T. | | Deposit date: | 2001-12-17 | | Release date: | 2002-02-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for the stabilization and inhibition of 2, 3-dihydroxybiphenyl 1,2-dioxygenase by t-butanol.
J.Biol.Chem., 273, 1998
|
|
1Y6O
 
 | | Crystal structure of disulfide engineered porcine pancreatic phospholipase A2 to group-X isozyme in complex with inhibitor MJ33 and phosphate ions | | Descriptor: | 1-HEXADECYL-3-TRIFLUOROETHYL-SN-GLYCERO-2-PHOSPHATE METHANE, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Yu, B.Z, Pan, Y.H, Jassen, M.J.W, Bahnson, B.J, Jain, M.K. | | Deposit date: | 2004-12-06 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and structural properties of disulfide engineered phospholipase a(2): insight into the role of disulfide bonding patterns.
Biochemistry, 44, 2005
|
|
1KOK
 
 | | Crystal Structure of Mesopone Cytochrome c Peroxidase (MpCcP) | | Descriptor: | Cytochrome c Peroxidase, FE(III)-(4-MESOPORPHYRINONE) | | Authors: | Bhaskar, B, Immoos, C.E, Cohen, M.S, Barrows, T.P, Farmer, P.J, Poulos, T.L. | | Deposit date: | 2001-12-20 | | Release date: | 2002-10-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mesopone cytochrome c peroxidase: functional model of heme oxygenated oxidases.
J.Inorg.Biochem., 91, 2002
|
|
6CZI
 
 | | Structure of a redesigned beta barrel, mFAP1, bound to DFHBI | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one, mFAP1 | | Authors: | Doyle, L.A, Stoddard, B.L. | | Deposit date: | 2018-04-09 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | De novo design of a fluorescence-activating beta-barrel.
Nature, 561, 2018
|
|
2IMN
 
 | |
