3T62
 
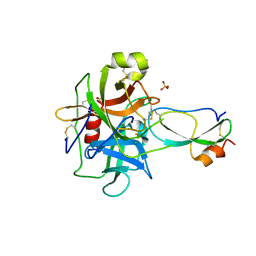 | | Crystal structure of recombinant Kunitz Type serine protease Inhibitor-1 from the Caribbean Sea anemone Stichodactyla helianthus in complex with bovine chymotrypsin | | Descriptor: | Chymotrypsinogen A, Kunitz-type proteinase inhibitor SHPI-1, SULFATE ION | | Authors: | Garcia-Fernandez, R, Dominguez, R, Oberthuer, D, Pons, T, Gonzalez-Gonzalez, Y, Chavez, M.A, Betzel, C, Redecke, L. | | Deposit date: | 2011-07-28 | | Release date: | 2012-08-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into chymotrypsin inhibition by the Kunitz-type inhibitor-1 from the marine invertebrate Stichodactyla helianthus
To be Published
|
|
3UOU
 
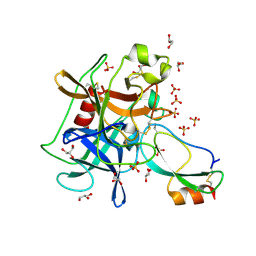 | | Crystal structure of the Kunitz-type protease inhibitor ShPI-1 Lys13Leu mutant in complex with pancreatic elastase | | Descriptor: | Chymotrypsin-like elastase family member 1, GLYCEROL, Kunitz-type proteinase inhibitor SHPI-1, ... | | Authors: | Garcia-Fernandez, R, Perbandt, M, Rehders, D, Gonzalez-Gonzalez, Y, Chavez, M.A, Betzel, C, Redecke, L. | | Deposit date: | 2011-11-17 | | Release date: | 2012-11-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional Structure of a Kunitz-type Inhibitor in Complex with an Elastase-like Enzyme.
J.Biol.Chem., 290, 2015
|
|
3M7Q
 
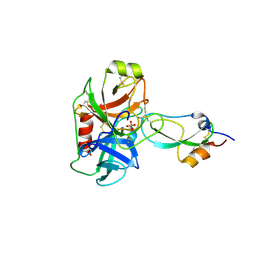 | | Crystal structure of recombinant Kunitz Type serine protease Inhibitor-1 from the Caribbean sea anemone stichodactyla helianthus in complex with bovine pancreatic trypsin | | Descriptor: | Cationic trypsin, Kunitz-type proteinase inhibitor SHPI-1, PHOSPHATE ION | | Authors: | Garcia-Fernandez, R, Redecke, L, Pons, T, Perbandt, M, Gil, D, Talavera, A, Gonzalez, Y, de los angeles Chavez, M, Betzel, C. | | Deposit date: | 2010-03-17 | | Release date: | 2011-03-16 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into serine protease inhibition by a marine invertebrate BPTI Kunitz-type inhibitor.
J.Struct.Biol., 180, 2012
|
|
3O1F
 
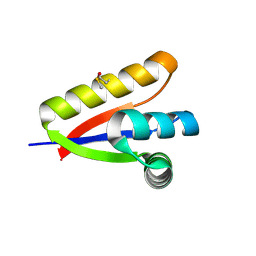 | | P1 crystal form of E. coli ClpS at 1.4 A resolution | | Descriptor: | ATP-dependent Clp protease adapter protein clpS | | Authors: | Roman-Hernandez, G, Hou, J.Y, Grant, R.A, Sauer, R.T, Baker, T.A. | | Deposit date: | 2010-07-21 | | Release date: | 2011-07-27 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The ClpS Adaptor Mediates Staged Delivery of N-End Rule Substrates to the AAA+ ClpAP Protease.
Mol.Cell, 43, 2011
|
|
3O2B
 
 | | E. coli ClpS in complex with a Phe N-end rule peptide | | Descriptor: | ATP-dependent Clp protease adaptor protein ClpS, CHLORIDE ION, Phe N-end rule peptide, ... | | Authors: | Roman-Hernandez, G, Grant, R.A, Sauer, R.T, Baker, T.A, de Regt, A. | | Deposit date: | 2010-07-22 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The ClpS adaptor mediates staged delivery of N-end rule substrates to the AAA+ ClpAP protease.
Mol.Cell, 43, 2011
|
|
3O2H
 
 | | E. coli ClpS in complex with a Leu N-end rule peptide | | Descriptor: | ATP-dependent Clp protease adaptor protein ClpS, DNA protection during starvation protein | | Authors: | Roman-Hernandez, G, Grant, R.A, Sauer, R.T, Baker, T.A, de Regt, A. | | Deposit date: | 2010-07-22 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The ClpS adaptor mediates staged delivery of N-end rule substrates to the AAA+ ClpAP protease.
Mol.Cell, 43, 2011
|
|
3O2O
 
 | | Structure of E. coli ClpS ring complex | | Descriptor: | ATP-dependent Clp protease adaptor protein ClpS | | Authors: | Roman-Hernandez, G, Grant, R.A, Sauer, R.T, Baker, T.A, de Regt, A. | | Deposit date: | 2010-07-22 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The ClpS adaptor mediates staged delivery of N-end rule substrates to the AAA+ ClpAP protease.
Mol.Cell, 43, 2011
|
|
3BB7
 
 | | Structure of Prevotella intermedia prointerpain A fragment 39-359 (mutant C154A) | | Descriptor: | interpain A | | Authors: | Mallorqui-Fernandez, N, Manandhar, S.P, Mallorqui-Fernandez, G, Uson, I, Wawrzonek, K, Kantyka, T, Sola, M, Thogersen, I.B, Enghild, J.J, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2007-11-09 | | Release date: | 2007-11-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A New Autocatalytic Activation Mechanism for Cysteine Proteases Revealed by Prevotella intermedia Interpain A
J.Biol.Chem., 283, 2008
|
|
3BBA
 
 | | Structure of active wild-type Prevotella intermedia interpain A cysteine protease | | Descriptor: | interpain A | | Authors: | Mallorqui-Fernandez, N, Manandhar, S.P, Mallorqui-Fernandez, G, Uson, I, Wawrzonek, K, Kantyka, T, Sola, M, Thogersen, I.B, Enghild, J.J, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2007-11-09 | | Release date: | 2007-11-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A New Autocatalytic Activation Mechanism for Cysteine Proteases Revealed by Prevotella intermedia Interpain A
J.Biol.Chem., 283, 2008
|
|
4JZ5
 
 | |
1RN7
 
 | | Structure of human cystatin D | | Descriptor: | Cystatin D | | Authors: | Alvarez-Fernandez, M, Liang, Y.H, Abrahamson, M, Su, X.D. | | Deposit date: | 2003-11-30 | | Release date: | 2004-05-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human cystatin D, a cysteine peptidase inhibitor with restricted inhibition profile.
J.Biol.Chem., 280, 2005
|
|
1ROA
 
 | | Structure of human cystatin D | | Descriptor: | Cystatin D | | Authors: | Alvarez-Fernandez, M, Liang, Y.H, Abrahamson, M, Su, X.D. | | Deposit date: | 2003-12-01 | | Release date: | 2004-05-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human cystatin D, a cysteine peptidase inhibitor with restricted inhibition profile.
J.Biol.Chem., 280, 2005
|
|
6IBQ
 
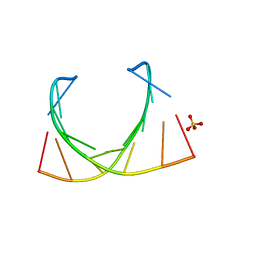 | | Structure of a nonameric RNA duplex at room temperature in ChipX microfluidic device | | Descriptor: | DNA/RNA (5'-R(*CP*GP*UP*GP*AP*UP*CP*G)-D(P*C)-3'), SULFATE ION | | Authors: | de Wijn, R, Olieric, V, Lorber, B, Sauter, C. | | Deposit date: | 2018-11-30 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A simple and versatile microfluidic device for efficient biomacromolecule crystallization and structural analysis by serial crystallography.
Iucrj, 6, 2019
|
|
3QSK
 
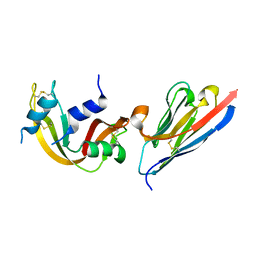 | | 5 Histidine Variant of the anti-RNase A VHH in Complex with RNAse A | | Descriptor: | Engineered 5 Histidine anti-RNase A Camelid VHH Antibody Domain Variant, Ribonuclease pancreatic | | Authors: | Murtaugh, M.L, Fanning, S.W, Sharma, T.M, Terry, A.M, Horn, J.R. | | Deposit date: | 2011-02-21 | | Release date: | 2011-08-10 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A combinatorial histidine scanning library approach to engineer highly pH-dependent protein switches.
Protein Sci., 20, 2011
|
|
3J6X
 
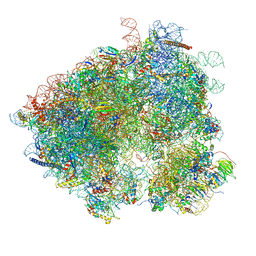 | | S. cerevisiae 80S ribosome bound with Taura syndrome virus (TSV) IRES, 5 degree rotation (Class II) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0, ... | | Authors: | Koh, C.S, Brilot, A.F, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2014-04-16 | | Release date: | 2014-06-11 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Taura syndrome virus IRES initiates translation by binding its tRNA-mRNA-like structural element in the ribosomal decoding center.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
9N2B
 
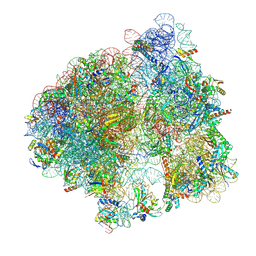 | | Impacts of ribosomal RNA sequence variation on gene expression and phenotype: Cryo-EM structure of the rrsB ribosome (BBB-70S) | | Descriptor: | 1,4-DIAMINOBUTANE, 16S ribosomal RNA (rRNA) from the rrnB operon, 23S ribosomal RNA (rRNA) from the rrnB operon, ... | | Authors: | Welfer, G.A, Brady, R.A, Natchiar, S.K, Watson, Z.L, Rundlet, E.J, Alejo, J.L, Singh, A.P, Mishra, N.K, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2025-01-28 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Impacts of ribosomal RNA sequence variation on gene expression and phenotype.
Philos.Trans.R.Soc.Lond.B Biol.Sci., 380, 2025
|
|
5D9A
 
 | | Influenza C Virus RNA-dependent RNA Polymerase - Space group P212121 | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA-directed RNA polymerase catalytic subunit | | Authors: | Hengrung, N, El Omari, K, Serna Martin, I, Vreede, F.T, Cusack, S, Rambo, R.P, Vonrhein, C, Bricogne, G, Stuart, D.I, Grimes, J.M, Fodor, E. | | Deposit date: | 2015-08-18 | | Release date: | 2015-10-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Crystal structure of the RNA-dependent RNA polymerase from influenza C virus.
Nature, 527, 2015
|
|
9N2C
 
 | | Impacts of ribosomal RNA sequence variation on gene expression and phenotype: Cryo-EM structure of the rrsH ribosome (HBB-70S) | | Descriptor: | 1,4-DIAMINOBUTANE, 16S ribosomal RNA (rRNA) from the rrnH operon, 23S ribosomal RNA (rRNA) from the rrnB operon, ... | | Authors: | Welfer, G.A, Brady, R.A, Natchiar, S.K, Watson, Z.L, Rundlet, E.J, Alejo, J.L, Singh, A.P, Mishra, N.K, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2025-01-28 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Impacts of ribosomal RNA sequence variation on gene expression and phenotype.
Philos.Trans.R.Soc.Lond.B Biol.Sci., 380, 2025
|
|
5XWY
 
 | | Electron cryo-microscopy structure of LbuCas13a-crRNA binary complex | | Descriptor: | A type VI-A CRISPR-Cas RNA-guided RNA ribonuclease, Cas13a, RNA (59-MER) | | Authors: | Zhang, X, Wang, Y, Ma, J, Liu, L, Li, X, Li, Z, You, L, Wang, J, Wang, M. | | Deposit date: | 2017-06-30 | | Release date: | 2017-09-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The Molecular Architecture for RNA-Guided RNA Cleavage by Cas13a.
Cell, 170, 2017
|
|
6TNA
 
 | | CRYSTAL STRUCTURE OF YEAST PHENYLALANINE T-RNA. I.CRYSTALLOGRAPHIC REFINEMENT | | Descriptor: | MAGNESIUM ION, TRNAPHE | | Authors: | Sussman, J.L, Holbrook, S.R, Warrant, R.W, Church, G.M, Kim, S.-H. | | Deposit date: | 1978-11-16 | | Release date: | 1979-01-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of yeast phenylalanine transfer RNA. I. Crystallographic refinement.
J.Mol.Biol., 123, 1978
|
|
1L1C
 
 | | Structure of the LicT Bacterial Antiterminator Protein in Complex with its RNA Target | | Descriptor: | Transcription antiterminator licT, licT mRNA antiterminator hairpin | | Authors: | Yang, Y, Declerck, N, Manival, X, Aymerich, S, Kochoyan, M. | | Deposit date: | 2002-02-15 | | Release date: | 2002-03-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the LicT-RNA antitermination complex: CAT clamping RAT.
EMBO J., 21, 2002
|
|
4V5Q
 
 | | The crystal structure of EF-Tu and G24A-tRNA-Trp bound to a near- cognate codon on the 70S ribosome | | Descriptor: | 16S RRNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Schmeing, T.M, Voorhees, R.M, Ramakrishnan, V. | | Deposit date: | 2010-12-07 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | How Mutations in tRNA Distant from the Anticodon Affect the Fidelity of Decoding.
Nat.Struct.Mol.Biol., 18, 2011
|
|
9J8G
 
 | | mouse zbp1 hybrid complex | | Descriptor: | DNA (5'-D(P*CP*AP*CP*GP*CP*A)-3'), RNA (5'-R(P*UP*GP*CP*GP*UP*G)-3'), Z-DNA-binding protein 1 | | Authors: | Gao, A.M, Zhoiu, C. | | Deposit date: | 2024-08-21 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | ZBP1 senses spliceosome stress through Z-RNA:DNA hybrid recognition.
Mol.Cell, 85, 2025
|
|
9J89
 
 | | zbp1 nucleic acid complex | | Descriptor: | DNA (5'-D(P*CP*AP*CP*GP*CP*A)-3'), MAGNESIUM ION, RNA (5'-R(P*UP*GP*CP*GP*UP*G)-3'), ... | | Authors: | Gao, A.M, Zhou, C. | | Deposit date: | 2024-08-20 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | ZBP1 senses spliceosome stress through Z-RNA:DNA hybrid recognition.
Mol.Cell, 85, 2025
|
|
4V5R
 
 | | The crystal structure of EF-Tu and Trp-tRNA-Trp bound to a cognate codon on the 70S ribosome. | | Descriptor: | 16S RRNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Schmeing, T.M, Voorhees, R.M, Ramakrishnan, V. | | Deposit date: | 2010-12-07 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | How Mutations in tRNA Distant from the Anticodon Affect the Fidelity of Decoding.
Nat.Struct.Mol.Biol., 18, 2011
|
|
