3S7J
 
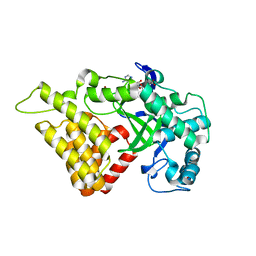 | |
3TZY
 
 | | Crystal structure of a fragment containing the acyltransferase domain of Pks13 from Mycobacterium tuberculosis in the palmitoylated form at 2.2 A | | Descriptor: | 12-mer peptide, GLYCEROL, PALMITIC ACID, ... | | Authors: | Bergeret, F, Pedelacq, J.D, Mourey, L, Bon, C. | | Deposit date: | 2011-09-28 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical and structural study of the atypical acyltransferase domain from the mycobacterial polyketide synthase pks13
J.Biol.Chem., 287, 2012
|
|
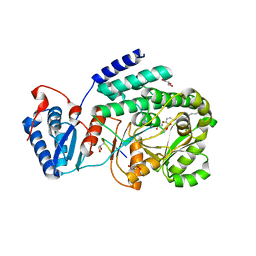
3TZW
 
 | | Crystal structure of a fragment containing the acyltransferase domain of Pks13 from Mycobacterium tuberculosis in the orthorhombic apoform at 2.6 A | | Descriptor: | 1,2-ETHANEDIOL, 12-mer peptide, Polyketide synthase PKS13, ... | | Authors: | Bergeret, F, Pedelacq, J.D, Mourey, L, Bon, C. | | Deposit date: | 2011-09-28 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Biochemical and structural study of the atypical acyltransferase domain from the mycobacterial polyketide synthase pks13
J.Biol.Chem., 287, 2012
|
|
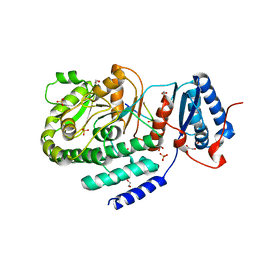
3TZX
 
 | | Crystal structure of a fragment containing the acyltransferase domain of Pks13 from Mycobacterium tuberculosis in tetragonal apo form at 2.3 A | | Descriptor: | 12-mer peptide, GLYCEROL, Polyketide synthase PKS13, ... | | Authors: | Bergeret, F, Pedelacq, J.D, Mourey, L, Bon, C. | | Deposit date: | 2011-09-28 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biochemical and structural study of the atypical acyltransferase domain from the mycobacterial polyketide synthase pks13
J.Biol.Chem., 287, 2012
|
|
7KJB
 
 | |
7KJA
 
 | |
7KJC
 
 | |
5TNC
 
 | |
5UQO
 
 | |
5UQU
 
 | |
5UQQ
 
 | |
5V60
 
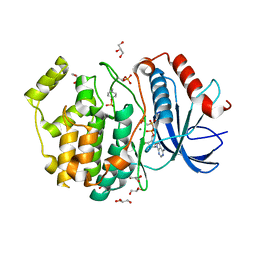 | | Phospho-ERK2 bound to AMP-PCP | | Descriptor: | GLYCEROL, MAGNESIUM ION, Mitogen-activated protein kinase 1, ... | | Authors: | Lechtenberg, B.C, Riedl, S.J. | | Deposit date: | 2017-03-15 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure-Guided Strategy for the Development of Potent Bivalent ERK Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
5V61
 
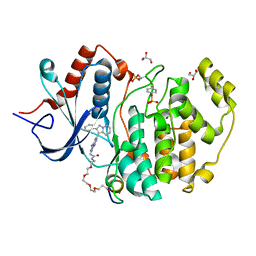 | | Phospho-ERK2 bound to bivalent inhibitor SBP2 | | Descriptor: | 2-oxo-6,9,12,15-tetraoxa-3-azaoctadecan-18-oic acid, 5-(2-PHENYLPYRAZOLO[1,5-A]PYRIDIN-3-YL)-1H-PYRAZOLO[3,4-C]PYRIDAZIN-3-AMINE, GLYCEROL, ... | | Authors: | Lechtenberg, B.C, Riedl, S.J. | | Deposit date: | 2017-03-15 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Strategy for the Development of Potent Bivalent ERK Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
5V62
 
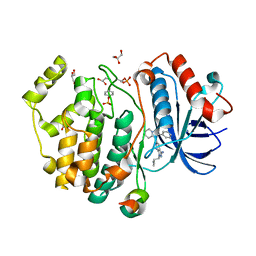 | | Phospho-ERK2 bound to bivalent inhibitor SBP3 | | Descriptor: | 5-(2-PHENYLPYRAZOLO[1,5-A]PYRIDIN-3-YL)-1H-PYRAZOLO[3,4-C]PYRIDAZIN-3-AMINE, GLYCEROL, Mitogen-activated protein kinase 1, ... | | Authors: | Lechtenberg, B.C, Riedl, S.J. | | Deposit date: | 2017-03-15 | | Release date: | 2017-07-26 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Strategy for the Development of Potent Bivalent ERK Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
5TVQ
 
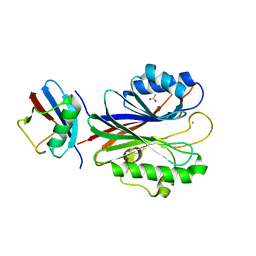 | |
5TVP
 
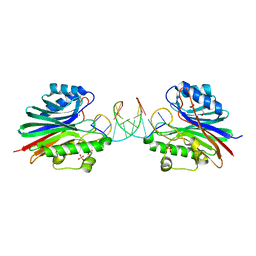 | |
5VAC
 
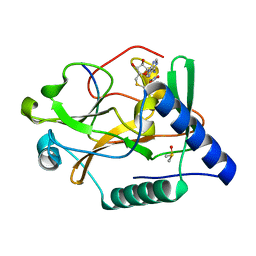 | | Crystal Structure of ATXR5 SET domain in complex with K36me3 histone H3 peptide | | Descriptor: | DIMETHYL SULFOXIDE, Histone H3.2, Probable Histone-lysine N-methyltransferase ATXR5, ... | | Authors: | Bergamin, E, Sarvan, S, Malette, J, Eram, M, Yeung, S, Mongeon, V, Joshi, M, Brunzelle, J.S, Michaels, S.D, Blais, A, Vedadi, M, Couture, J.F. | | Deposit date: | 2017-03-24 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Molecular basis for the methylation specificity of ATXR5 for histone H3.
Nucleic Acids Res., 45, 2017
|
|
5VA6
 
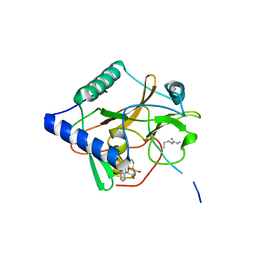 | | CRYSTAL STRUCTURE OF ATXR5 IN COMPLEX WITH HISTONE H3.1 MONO-METHYLATED ON R26 | | Descriptor: | Histone H3.1, Probable Histone-lysine N-methyltransferase ATXR5, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Bergamin, E, Sarvan, S, Malette, J, Eram, M, Yeung, S, Mongeon, V, Joshi, M, Brunzelle, J.S, Michaels, S.D, Blais, A, Vedadi, M, Couture, J.-F. | | Deposit date: | 2017-03-24 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for the methylation specificity of ATXR5 for histone H3.
Nucleic Acids Res., 45, 2017
|
|
5VAH
 
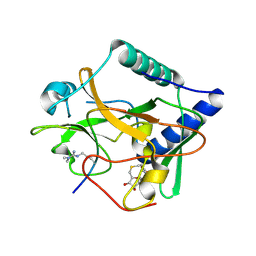 | | Crystal structure of ATXR5 SET domain in complex with histone H3 di-methylated on R26 | | Descriptor: | Histone H3.2, Probable Histone-lysine N-methyltransferase ATXR5, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Bergamin, E, Sarvan, S, Malette, J, Eram, M, Yeung, S, Mongeon, V, Joshi, M, Brunzelle, J.S, Michaels, S.D, Blais, A, Vedadi, M, Couture, J.-F. | | Deposit date: | 2017-03-26 | | Release date: | 2017-04-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for the methylation specificity of ATXR5 for histone H3.
Nucleic Acids Res., 45, 2017
|
|
5VAB
 
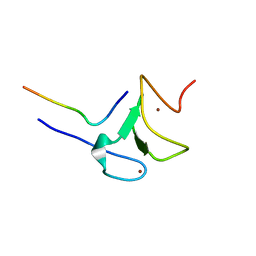 | | Crystal structure of ATXR5 PHD domain in complex with histone H3 | | Descriptor: | ATXR5 PHD domain, Histone H3 peptide, ZINC ION | | Authors: | Bergamin, E, Sarvan, S, Malette, J, Eram, M, Yeung, S, Mongeon, V, Joshi, M, Brunzelle, J.S, Michaels, S.D, Blais, A, Vedadi, M, Couture, J.-F. | | Deposit date: | 2017-03-24 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Molecular basis for the methylation specificity of ATXR5 for histone H3.
Nucleic Acids Res., 45, 2017
|
|
1LGL
 
 | | Solution structure of HERG-specific scorpion toxin BeKm-1 | | Descriptor: | BeKm-1 toxin | | Authors: | Korolokova, Y.V, Bocharov, E.V, Angelo, K, Maslennikov, I.V, Grinenko, O.V, Lipkin, A.V, Nosireva, E.D, Pluzhnikov, K.A, Olesen, S.-P, Arseniev, A.S, Grishin, E.V. | | Deposit date: | 2002-04-16 | | Release date: | 2002-11-20 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | New binding site on common molecular scaffold provides HERG channel specificity of scorpion toxin BeKm-1.
J.Biol.Chem., 277, 2002
|
|
2N7G
 
 | |
2L1M
 
 | |
1J5J
 
 | | Solution structure of HERG-specific scorpion toxin BeKm-1 | | Descriptor: | BeKm-1 toxin | | Authors: | Korolokova, Y.V, Bocharov, E.V, Angelo, K, Maslennikov, I.V, Grinenko, O.V, Lipkin, A.V, Nosireva, E.D, Pluzhnikov, K.A, Olesen, S.-P, Arseniev, A.S, Grishin, E.V. | | Deposit date: | 2002-04-16 | | Release date: | 2002-11-20 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | New binding site on common molecular scaffold provides HERG channel specificity of scorpion toxin BeKm-1.
J.Biol.Chem., 277, 2002
|
|
2L4R
 
 | | NMR solution structure of the N-terminal PAS domain of hERG | | Descriptor: | Potassium voltage-gated channel subfamily H member 2 | | Authors: | Gayen, N, Li, Q, Chen, A.S, Huang, Q, Raida, M, Kang, C. | | Deposit date: | 2010-10-13 | | Release date: | 2010-12-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the N-terminal domain of hERG and its interaction with the S4-S5 linker.
Biochem.Biophys.Res.Commun., 403, 2010
|
|
