2MQK
 
 | |
3H87
 
 | | Rv0301 Rv0300 Toxin Antitoxin Complex from Mycobacterium tuberculosis | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, IMIDAZOLE, ... | | Authors: | Min, A, Sawaya, M.R, Cascio, D, Eisenberg, D, Integrated Center for Structure and Function Innovation (ISFI) | | Deposit date: | 2009-04-28 | | Release date: | 2009-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | The crystal structure of the Rv0301-Rv0300 VapBC-3 toxin-antitoxin complex from M. tuberculosis reveals a Mg(2+) ion in the active site and a putative RNA-binding site.
Protein Sci., 21, 2012
|
|
5VA6
 
 | | CRYSTAL STRUCTURE OF ATXR5 IN COMPLEX WITH HISTONE H3.1 MONO-METHYLATED ON R26 | | Descriptor: | Histone H3.1, Probable Histone-lysine N-methyltransferase ATXR5, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Bergamin, E, Sarvan, S, Malette, J, Eram, M, Yeung, S, Mongeon, V, Joshi, M, Brunzelle, J.S, Michaels, S.D, Blais, A, Vedadi, M, Couture, J.-F. | | Deposit date: | 2017-03-24 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for the methylation specificity of ATXR5 for histone H3.
Nucleic Acids Res., 45, 2017
|
|
5W0V
 
 | | Crystal structure of full-length Kluyveromyces lactis Kap123 with histone H4 1-34 | | Descriptor: | Histone H4 1-34, Kap123 | | Authors: | An, S, Yoon, J, Song, J.-J, Cho, U.-S. | | Deposit date: | 2017-05-31 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.821 Å) | | Cite: | Structure-based nuclear import mechanism of histones H3 and H4 mediated by Kap123.
Elife, 6, 2017
|
|
2V89
 
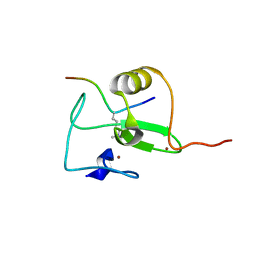 | |
2KQC
 
 | | Second PBZ domain of human APLF protein | | Descriptor: | Aprataxin and PNK-like factor, ZINC ION | | Authors: | Neuhaus, D, Eustermann, S, Brockmann, C, Yang, J. | | Deposit date: | 2009-11-04 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the two PBZ domains from human APLF and their interaction with poly(ADP-ribose).
Nat.Struct.Mol.Biol., 17, 2010
|
|
2KQB
 
 | | First PBZ domain of human APLF protein | | Descriptor: | Aprataxin and PNK-like factor, ZINC ION | | Authors: | Neuhaus, D, Eustermann, S, Brockmann, C, Yang, J. | | Deposit date: | 2009-11-04 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the two PBZ domains from human APLF and their interaction with poly(ADP-ribose).
Nat.Struct.Mol.Biol., 17, 2010
|
|
1QCK
 
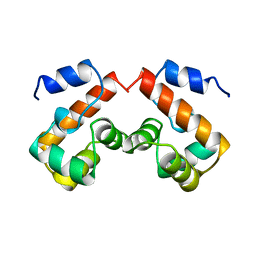 | |
2CSD
 
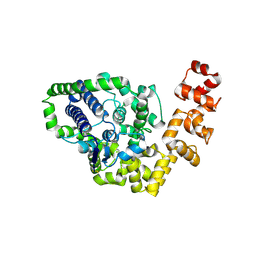 | |
1IPI
 
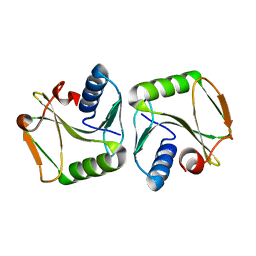 | |
4DM6
 
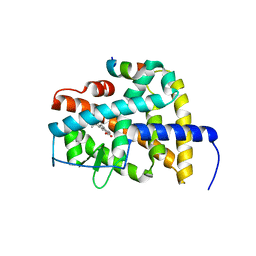 | | Crystal structure of RARb LBD homodimer in complex with TTNPB | | Descriptor: | 4-[(1E)-2-(5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRONAPHTHALEN-2-YL)PROP-1-ENYL]BENZOIC ACID, Nuclear receptor coactivator 1, Retinoic acid receptor beta | | Authors: | Osz, J, Brelivet, Y, Peluso-Iltis, C, Cura, V, Eiler, S, Ruff, M, Bourguet, W, Rochel, N, Moras, D. | | Deposit date: | 2012-02-07 | | Release date: | 2012-03-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for a molecular allosteric control mechanism of cofactor binding to nuclear receptors.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2KQD
 
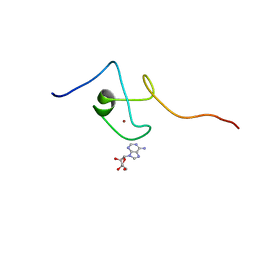 | | First PBZ domain of human APLF protein in complex with ribofuranosyladenosine | | Descriptor: | ADENOSINE, Aprataxin and PNK-like factor, ZINC ION, ... | | Authors: | Neuhaus, D, Eustermann, S, Brockmann, C, Yang, J. | | Deposit date: | 2009-11-04 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the two PBZ domains from human APLF and their interaction with poly(ADP-ribose).
Nat.Struct.Mol.Biol., 17, 2010
|
|
1U3C
 
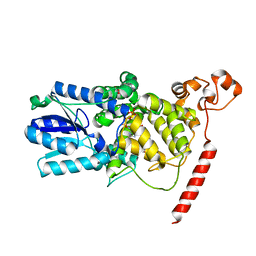 | | Crystal Structure of the PHR domain of Cryptochrome 1 from Arabidopsis thaliana | | Descriptor: | CHLORIDE ION, Cryptochrome 1 apoprotein, ETHYL DIMETHYL AMMONIO PROPANE SULFONATE, ... | | Authors: | Brautigam, C.A, Smith, B.S, Ma, Z, Palnitkar, M, Tomchick, D.R, Machius, M, Deisenhofer, J. | | Deposit date: | 2004-07-21 | | Release date: | 2004-08-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the photolyase-like domain of cryptochrome 1 from Arabidopsis thaliana.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2N25
 
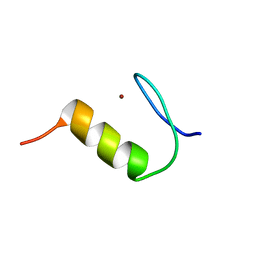 | | Solution structure of Miz-1 zinc finger 2 | | Descriptor: | ZINC ION, Zinc finger and BTB domain-containing protein 17 | | Authors: | Bedard, M, Lavigne, P. | | Deposit date: | 2015-04-28 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 13th C2H2 Zinc Finger of Miz-1.
Biochem.Biophys.Res.Commun., 473, 2016
|
|
2KQE
 
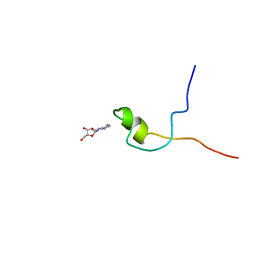 | | Second PBZ domain of human APLF protein in complex with ribofuranosyladenosine | | Descriptor: | ADENOSINE, Aprataxin and PNK-like factor, ZINC ION, ... | | Authors: | Neuhaus, D, Eustermann, S, Brockmann, C, Yang, J. | | Deposit date: | 2009-11-04 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the two PBZ domains from human APLF and their interaction with poly(ADP-ribose).
Nat.Struct.Mol.Biol., 17, 2010
|
|
4V7N
 
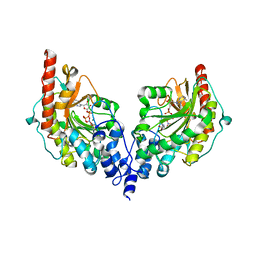 | | Glycocyamine kinase, beta-beta homodimer from marine worm Namalycastis sp., with transition state analog Mg(II)-ADP-NO3-glycocyamine. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANIDINO ACETATE, Glycocyamine kinase beta chain, ... | | Authors: | Lim, K, Pullalarevu, S, Herzberg, O. | | Deposit date: | 2009-12-15 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the mechanism and substrate specificity of glycocyamine kinase, a phosphagen kinase family member.
Biochemistry, 49, 2010
|
|
6V0L
 
 | | PDGFR-b Promoter Forms a G-Vacancy Quadruplex that Can be Complemented by dGMP: Molecular Structure and Recognition of Guanine Derivatives and Metabolites | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, DNA (5'-D(*(3D1)P*AP*GP*GP*GP*AP*GP*GP*GP*CP*GP*GP*CP*GP*GP*GP*AP*CP*A)-3') | | Authors: | Wang, K.B, Dickerhoff, J, Wu, G, Yang, D. | | Deposit date: | 2019-11-18 | | Release date: | 2020-03-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | PDGFR-beta Promoter Forms a Vacancy G-Quadruplex that Can Be Filled in by dGMP: Solution Structure and Molecular Recognition of Guanine Metabolites and Drugs.
J.Am.Chem.Soc., 142, 2020
|
|
3HVT
 
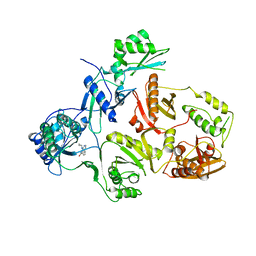 | | STRUCTURAL BASIS OF ASYMMETRY IN THE HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 REVERSE TRANSCRIPTASE HETERODIMER | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P51), HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P66) | | Authors: | Steitz, T.A, Smerdon, S.J, Jaeger, J, Wang, J, Kohlstaedt, L.A, Chirino, A.J, Friedman, J.M, Rice, P.A. | | Deposit date: | 1994-07-25 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the binding site for nonnucleoside inhibitors of the reverse transcriptase of human immunodeficiency virus type 1.
Proc.Natl.Acad.Sci.Usa, 91, 1994
|
|
6LQP
 
 | |
7DG5
 
 | |
1AO4
 
 | | COBALT(III)-PEPLOMYCIN COMPLEX DETERMINED BY NMR STUDIES | | Descriptor: | 3-O-carbamoyl-alpha-D-mannopyranose-(1-2)-alpha-L-gulopyranose, AGLYCON OF PEPLOMYCIN, COBALT (III) ION, ... | | Authors: | Caceres-Cortes, J, Sugiyama, H, Ikudome, K, Saito, I, Wang, A.H.-J. | | Deposit date: | 1997-07-16 | | Release date: | 1999-07-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structures of cobalt(III)-pepleomycin and cobalt(III)-deglycopepleomycin (green forms) determined by NMR studies.
Eur.J.Biochem., 244, 1997
|
|
3HJH
 
 | | A rigid N-terminal clamp restrains the motor domains of the bacterial transcription-repair coupling factor | | Descriptor: | COBALT (II) ION, Transcription-repair-coupling factor | | Authors: | Murphy, M, Gong, P, Ralto, K, Manelyte, L, Savery, N, Theis, K. | | Deposit date: | 2009-05-21 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | An N-terminal clamp restrains the motor domains of the bacterial transcription-repair coupling factor Mfd.
Nucleic Acids Res., 37, 2009
|
|
6LQU
 
 | |
1FTA
 
 | | FRUCTOSE-1,6-BISPHOSPHATASE(D-FRUCTOSE-1,6-BISPHOSPHATE, 1-PHOSPHOHYDROLASE) (E.C.3.1.3.11) COMPLEXED WITH THE ALLOSTERIC INHIBITOR AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, FRUCTOSE-1,6-BISPHOSPHATASE | | Authors: | Zhang, Y, Liang, J.-Y, Huang, S, Lipscomb, W.N. | | Deposit date: | 1993-09-27 | | Release date: | 1995-11-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The allosteric site of human liver fructose-1,6-bisphosphatase. Analysis of six AMP site mutants based on the crystal structure.
J.Biol.Chem., 269, 1994
|
|
7KWZ
 
 | | TDP-43 LCD amyloid fibrils | | Descriptor: | Isoform 2 of TAR DNA-binding protein 43 | | Authors: | Li, Q, Babinchak, W.M, Surewicz, W.K. | | Deposit date: | 2020-12-02 | | Release date: | 2021-02-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of amyloid fibrils formed by the entire low complexity domain of TDP-43.
Nat Commun, 12, 2021
|
|
