4PVD
 
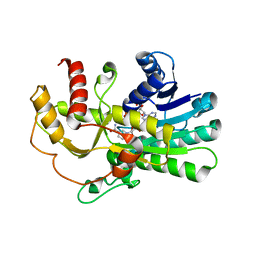 | | Crystal structure of yeast methylglyoxal/isovaleraldehyde reductase Gre2 complexed with NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH-dependent methylglyoxal reductase GRE2 | | Authors: | Guo, P.C, Bao, Z.Z, Li, W.F, Zhou, C.Z. | | Deposit date: | 2014-03-17 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the cofactor-assisted substrate recognition of yeast methylglyoxal/isovaleraldehyde reductase Gre2
Biochim.Biophys.Acta, 1844, 2014
|
|
7ZU6
 
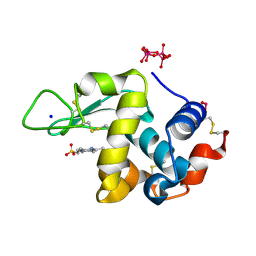 | | Polyoxidovanadate interaction with proteins: crystal structure of lysozyme bound to tetra-vanadate ion (structure 1) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CYCLO-TETRAMETAVANADATE, Lysozyme, ... | | Authors: | Tito, G, Merlino, A, Ferraro, G. | | Deposit date: | 2022-05-11 | | Release date: | 2023-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.183 Å) | | Cite: | Stabilization and Binding of [V 4 O 12 ] 4- and Unprecedented [V 20 O 54 (NO 3 )] n- to Lysozyme upon Loss of Ligands and Oxidation of the Potential Drug V IV O(acetylacetonato) 2.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
1ASZ
 
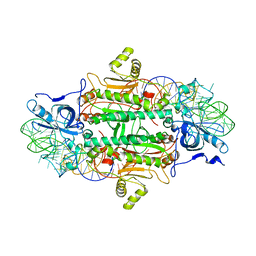 | | THE ACTIVE SITE OF YEAST ASPARTYL-TRNA SYNTHETASE: STRUCTURAL AND FUNCTIONAL ASPECTS OF THE AMINOACYLATION REACTION | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ASPARTYL-tRNA SYNTHETASE, T-RNA (75-MER) | | Authors: | Cavarelli, J, Rees, B, Thierry, J.C, Moras, D. | | Deposit date: | 1995-01-19 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation reaction.
EMBO J., 13, 1994
|
|
4PWV
 
 | |
4PXE
 
 | | The crystal structure of AtUAH in complex with glyoxylate | | Descriptor: | GLYOXYLIC ACID, MANGANESE (II) ION, Ureidoglycolate hydrolase | | Authors: | Shin, I, Rhee, S. | | Deposit date: | 2014-03-23 | | Release date: | 2014-07-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Structural insights into the substrate specificity of (s)-ureidoglycolate amidohydrolase and its comparison with allantoate amidohydrolase.
J.Mol.Biol., 426, 2014
|
|
6B9T
 
 | |
1AO6
 
 | |
5LWO
 
 | | Structure of Spin-labelled T4 lysozyme mutant L115C-R119C-R1 at 100K | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Loll, B, Consentius, P, Gohlke, U, Mueller, R, Kaupp, M, Heinemann, U, Wahl, M.C, Risse, T. | | Deposit date: | 2016-09-18 | | Release date: | 2017-03-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.183 Å) | | Cite: | Internal Dynamics of the 3-Pyrroline-N-Oxide Ring in Spin-Labeled Proteins.
J Phys Chem Lett, 8, 2017
|
|
5LTE
 
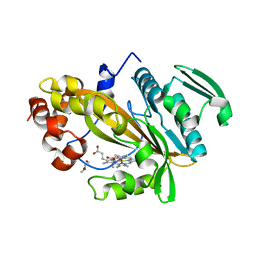 | |
5LUM
 
 | | Alpha-crystallin domain of human HSPB6 patched with its N-terminal peptide | | Descriptor: | Heat shock protein beta-6, SULFATE ION | | Authors: | Sluchanko, N.N, Beelen, S, Kulikova, A.A, Weeks, S.D, Antson, A.A, Gusev, N.B, Strelkov, S.V. | | Deposit date: | 2016-09-09 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for the Interaction of a Human Small Heat Shock Protein with the 14-3-3 Universal Signaling Regulator.
Structure, 25, 2017
|
|
5LUS
 
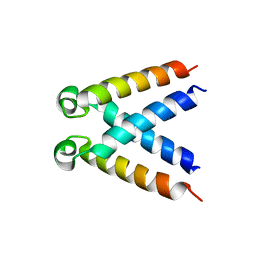 | | Structures of DHBN domain of Pelecanus crispus BLM helicase | | Descriptor: | BLM helicase | | Authors: | Shi, J, Chen, W.-F, Zhang, B, Fan, S.-H, Ai, X, Liu, N.-N, Rety, S, Xi, X.-G. | | Deposit date: | 2016-09-09 | | Release date: | 2017-03-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.433 Å) | | Cite: | A helical bundle in the N-terminal domain of the BLM helicase mediates dimer and potentially hexamer formation.
J. Biol. Chem., 292, 2017
|
|
1IJ8
 
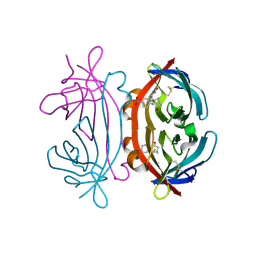 | | CRYSTAL STRUCTURE OF LITE AVIDIN-BNI COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-(2-OXO-HEXAHYDRO-THIENO[3,4-D]IMIDAZOL-6-YL)-PENTANOIC ACID (4-NITRO-PHENYL)-AMIDE, AVIDIN | | Authors: | Livnah, O, Huberman, T. | | Deposit date: | 2001-04-25 | | Release date: | 2001-09-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Chicken avidin exhibits pseudo-catalytic properties. Biochemical, structural, and electrostatic consequences.
J.Biol.Chem., 276, 2001
|
|
7ZWA
 
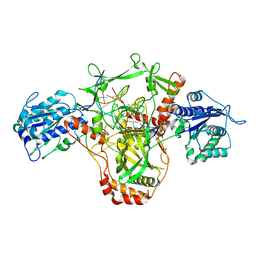 | | CryoEM structure of Ku heterodimer bound to DNA and PAXX | | Descriptor: | DNA (5'-D(P*CP*GP*AP*TP*AP*TP*CP*TP*AP*GP*AP*GP*GP*GP*AP*TP*C)-3'), DNA (5'-D(P*GP*AP*TP*CP*CP*CP*TP*CP*TP*AP*GP*AP*TP*AP*T)-3'), PHOSPHATE ION, ... | | Authors: | Hardwick, S.W, Kefala-Stavridi, A, Chirgadze, D.Y, Blundell, T.L, Chaplin, A.K. | | Deposit date: | 2022-05-19 | | Release date: | 2023-05-31 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | PAXX binding to the NHEJ machinery explains functional redundancy with XLF.
Sci Adv, 9, 2023
|
|
8DCS
 
 | | Cryo-EM structure of cyanopindolol-bound beta1-adrenergic receptor in complex with heterotrimeric Gs-protein | | Descriptor: | 4-{[(2S)-3-(tert-butylamino)-2-hydroxypropyl]oxy}-3H-indole-2-carbonitrile, Endolysin,Endolysin,Beta-1 adrenergic receptor chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Su, M, Paknejad, N, Hite, R.K, Huang, X.Y. | | Deposit date: | 2022-06-17 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structures of beta 1 -adrenergic receptor in complex with Gs and ligands of different efficacies.
Nat Commun, 13, 2022
|
|
7ZLT
 
 | |
6B5H
 
 | |
3WE4
 
 | |
4Q3S
 
 | | Crystal structure of Schistosoma mansoni arginase in complex with inhibitor ABHPE | | Descriptor: | Arginase, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Hai, Y, Edwards, J.E, Van Zandt, M.C, Hoffmann, K.F, Christianson, D.W. | | Deposit date: | 2014-04-12 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal Structure of Schistosoma mansoni Arginase, a Potential Drug Target for the Treatment of Schistosomiasis.
Biochemistry, 53, 2014
|
|
5LDR
 
 | | Crystal structure of a cold-adapted dimeric beta-D-galactosidase from Paracoccus sp. 32d strain in complex with galactose | | Descriptor: | ACETATE ION, Beta-D-galactosidase, CHLORIDE ION, ... | | Authors: | Rutkiewicz-Krotewicz, M, Bujacz, A, Pietrzyk, A.J, Sekula, B, Bujacz, G. | | Deposit date: | 2016-06-27 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural studies of a cold-adapted dimeric beta-D-galactosidase from Paracoccus sp. 32d.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
3WTM
 
 | | Crystal Structure of Lymnaea stagnalis Acetylcholine-Binding Protein Q55R Mutant Complexed with Nitromethylene Analogue of Imidacloprid | | Descriptor: | 2-chloro-5-{[(2E)-2-(nitromethylidene)imidazolidin-1-yl]methyl}pyridine, Acetylcholine-binding protein | | Authors: | Okajima, T, Ihara, M, Yamashita, A, Oda, T, Matsuda, K. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-04 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Studies on an acetylcholine binding protein identify a basic residue in loop G on the beta 1 strand as a new structural determinant of neonicotinoid actions
Mol.Pharmacol., 86, 2014
|
|
6B6L
 
 | | The crystal structure of glycosyl hydrolase family 2 (GH2) member from Bacteroides cellulosilyticus DSM 14838 | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Glycosyl hydrolase family 2, ... | | Authors: | Tan, K, Joachimiak, G, Nocek, B, Enddres, M, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-10-02 | | Release date: | 2017-10-11 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of glycosyl hydrolase family 2 (GH2) member from Bacteroides cellulosilyticus DSM 14838
To Be Published
|
|
8A9Q
 
 | | Computational design of stable mammalian serum albumins for bacterial expression | | Descriptor: | Albumin, LAURIC ACID, MYRISTIC ACID, ... | | Authors: | Khersonsky, O, Dym, O, Fleishman, J.S. | | Deposit date: | 2022-06-29 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Stable Mammalian Serum Albumins Designed for Bacterial Expression.
J.Mol.Biol., 435, 2023
|
|
5LZU
 
 | | Structure of the mammalian ribosomal termination complex with accommodated eRF1 | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Shao, S, Murray, J, Brown, A, Taunton, J, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2016-10-02 | | Release date: | 2016-11-30 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Decoding Mammalian Ribosome-mRNA States by Translational GTPase Complexes.
Cell, 167, 2016
|
|
5M1Z
 
 | |
8DCR
 
 | | Cryo-EM structure of dobutamine-bound beta1-adrenergic receptor in complex with heterotrimeric Gs-protein | | Descriptor: | DOBUTAMINE, Endolysin,Endolysin,Beta-1 adrenergic receptor chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Su, M, Paknejad, N, Hite, R.K, Huang, X.Y. | | Deposit date: | 2022-06-17 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures of beta 1 -adrenergic receptor in complex with Gs and ligands of different efficacies.
Nat Commun, 13, 2022
|
|
