2VS6
 
 | | K173A, R174A, K177A-trichosanthin | | Descriptor: | RIBOSOME-INACTIVATING PROTEIN ALPHA-TRICHOSANTHIN | | Authors: | Too, P.H, Ma, M.K, Mak, A.N, Tung, C.K, Zhu, G, Au, S.W, Wong, K.B, Shaw, P.C, Ng, A. | | Deposit date: | 2008-04-21 | | Release date: | 2008-12-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The C-Terminal Fragment of the Ribosomal P Protein Complexed to Trichosanthin Reveals the Interaction between the Ribosome-Inactivating Protein and the Ribosome.
Nucleic Acids Res., 37, 2009
|
|
8TJF
 
 | | monovalent bispecific IgG antibodies through novel electrostatic steering mutations at the CH1-CL interface | | Descriptor: | Fab Lambda light chain, IgG1 Fab heavy chain | | Authors: | Oganesyan, V.Y, van Dyk, N, Mazor, Y, Chiang, C. | | Deposit date: | 2023-07-21 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Robust production of monovalent bispecific IgG antibodies through novel electrostatic steering mutations at the C H 1-C lambda interface.
Mabs, 15, 2023
|
|
3PSB
 
 | | Furo[2,3-c]pyridine-based Indanone Oximes as Potent and Selective B-Raf Inhibitors | | Descriptor: | B-RAF PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE, ethyl 3-{[1-(hydroxyamino)-2H-inden-5-yl]amino}thieno[2,3-c]pyridine-2-carboxylate | | Authors: | Morales, T, Vigers, G.P.A, Brandhuber, B.J. | | Deposit date: | 2010-12-01 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The Discovery of furo[2,3-c]pyridine-based indanone oximes as potent and selective B-Raf inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2WJM
 
 | | Lipidic sponge phase crystal structure of the photosynthetic reaction centre from Blastochloris viridis (low dose) | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Woehri, A.B, Wahlgren, W.Y, Malmerberg, E, Johansson, L.C, Neutze, R, Katona, G. | | Deposit date: | 2009-05-27 | | Release date: | 2009-09-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Lipidic sponge phase crystal structure of a photosynthetic reaction center reveals lipids on the protein surface.
Biochemistry, 48, 2009
|
|
3BI6
 
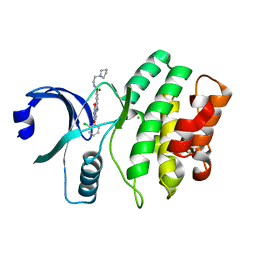 | | Wee1 kinase complex with inhibitor PD352396 | | Descriptor: | 4-(2-chlorophenyl)-9-hydroxy-6-methyl-1,3-dioxo-N-(2-pyrrolidin-1-ylethyl)pyrrolo[3,4-g]carbazole-8-carboxamide, CHLORIDE ION, Wee1-like protein kinase | | Authors: | Squire, C.J, Dickson, J.M, Ivanovic, I, Baker, E.N. | | Deposit date: | 2007-11-29 | | Release date: | 2007-12-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis and structure-activity relationships of soluble 8-substituted 4-(2-chlorophenyl)-9-hydroxypyrrolo[3,4-c]carbazole-1,3(2H,6H)-diones as inhibitors of the Wee1 and Chk1 checkpoint kinases.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3PUJ
 
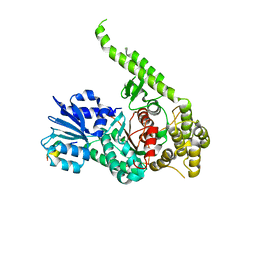 | | Crystal structure of the MUNC18-1 and SYNTAXIN4 N-Peptide complex | | Descriptor: | Syntaxin-4 N-terminal peptide, Syntaxin-binding protein 1 | | Authors: | Hu, S.-H, Christie, M.P, Saez, N.J, Latham, C.F, Jarrott, R, Lua, L.H.L, Collins, B.M, Martin, J.L. | | Deposit date: | 2010-12-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.313 Å) | | Cite: | Possible roles for Munc18-1 domain 3a and Syntaxin1 N-peptide and C-terminal anchor in SNARE complex formation
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1CYI
 
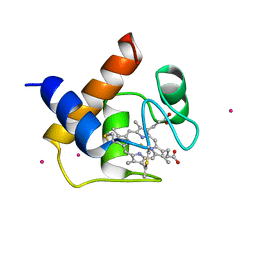 | | CYTOCHROME C6 | | Descriptor: | CADMIUM ION, CYTOCHROME C6, HEME C | | Authors: | Kerfeld, C.A, Yeates, T.O. | | Deposit date: | 1995-05-09 | | Release date: | 1996-01-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of chloroplast cytochrome c6 at 1.9 A resolution: evidence for functional oligomerization.
J.Mol.Biol., 250, 1995
|
|
5EDH
 
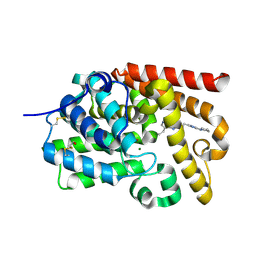 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH n4c(C)n1c(nc(n1)CCc2nc(nn2C)N3CCCC3)c(c4)CC, micromolar IC50=0.0037753 | | Descriptor: | 8-ethyl-5-methyl-2-[2-(2-methyl-5-pyrrolidin-1-yl-1,2,4-triazol-3-yl)ethyl]-[1,2,4]triazolo[1,5-c]pyrimidine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Rudolph, M.G. | | Deposit date: | 2015-10-21 | | Release date: | 2016-03-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A Real-World Perspective on Molecular Design.
J.Med.Chem., 59, 2016
|
|
3LV3
 
 | | Crystal structure of HLA-B*2705 complexed with a peptide derived from the human voltage-dependent calcium channel alpha1 subunit (residues 513-521) | | Descriptor: | 9-meric peptide from Voltage-dependent L-type calcium channel subunit alpha-1D, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Loll, B, Rueckert, C, Saenger, W, Uchanska-Ziegler, B, Ziegler, A. | | Deposit date: | 2010-02-19 | | Release date: | 2010-11-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Loss of recognition by cross-reactive T cells and its relation to a C-terminus-induced conformational reorientation of an HLA-B*2705-bound peptide.
Protein Sci., 20, 2011
|
|
5TBD
 
 | | Crystal Structure of anti-MSP2 Fv fragment (mAb4D11) in complex with 3D7-MSP2 215-222 | | Descriptor: | Fv fragment (mAb4D1) heavy chain, Merozoite surface protein 2 | | Authors: | Seow, J, Morales, R.A.V, MacRaild, C.A, Bankala, K, Drinkwater, N, Dingjan, T, Jaipuria, G, Wilde, K, Anders, R.F, Atreya, H.S, Christ, D, McGowan, S, Norton, R.S. | | Deposit date: | 2016-09-12 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Characterisation of a Key Epitope in the Conserved C-Terminal Domain of the Malaria Vaccine Candidate MSP2.
J. Mol. Biol., 429, 2017
|
|
2FK9
 
 | | Human protein kinase C, eta | | Descriptor: | protein kinase C, eta type | | Authors: | Walker, J.R, Littler, D.R, Finerty Jr, P.J, MacKenzie, F, Newman, E.M, Weigelt, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-01-04 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of human protein kinase C eta (PKCeta) C2 domain and identification of phosphorylation sites.
Biochem.Biophys.Res.Commun., 349, 2006
|
|
1I5U
 
 | | SOLUTION STRUCTURE OF CYTOCHROME B5 TRIPLE MUTANT (E48A/E56A/D60A) | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Qian, C, Yao, Y, Tang, W, Wang, J, Zhongxian, H. | | Deposit date: | 2001-02-28 | | Release date: | 2001-03-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Effects of charged amino-acid mutation on the solution structure of cytochrome b(5) and binding between cytochrome b(5) and cytochrome c.
Protein Sci., 10, 2001
|
|
5IWY
 
 | | Crystal structure of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase from Bacillus subtitis complexed with CMP and Mg2+ | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, CYTIDINE-5'-MONOPHOSPHATE, MAGNESIUM ION | | Authors: | Liu, Z.C, Jin, Y, Wang, G.G. | | Deposit date: | 2016-03-23 | | Release date: | 2017-03-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structure of IspF from Bacillus subtilis and absence of protein complex assembly among IspD/IspE/IspF enzymes in the MEP pathway.
Biosci. Rep., 2018
|
|
4OBX
 
 | | Crystal structure of yeast Coq5 in the apo form | | Descriptor: | 2-methoxy-6-polyprenyl-1,4-benzoquinol methylase, mitochondrial, TRIS(HYDROXYETHYL)AMINOMETHANE | | Authors: | Dai, Y.N, Zhou, K, Cao, D.D, Jiang, Y.L, Meng, F, Chi, C.B, Ren, Y.M, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2014-01-07 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures and catalytic mechanism of the C-methyltransferase Coq5 provide insights into a key step of the yeast coenzyme Q synthesis pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5EQQ
 
 | | Crystal structure of HCV NS3/4A WT protease in complex with 5172-Linear (MK-5172 linear analogue) | | Descriptor: | NS3 protease, SULFATE ION, ZINC ION, ... | | Authors: | Soumana, D, Yilmaz, N.K, Ali, A, Prachanronarong, K.L, Aydin, C, Schiffer, C.A. | | Deposit date: | 2015-11-13 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Thermodynamic Effects of Macrocyclization in HCV NS3/4A Inhibitor MK-5172.
Acs Chem.Biol., 11, 2016
|
|
4NM2
 
 | | Structure of human DNA polymerase beta complexed with a nicked DNA containing a 8BrG-G at N-1 position and G-C at N position | | Descriptor: | 5'-D(*CP*CP*GP*AP*CP*GP*(BGM)P*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*GP*C)-3', 5'-D(P*GP*TP*CP*GP*G)-3', ... | | Authors: | Koag, M.-C, Min, K, Monzingo, A.F, Lee, S. | | Deposit date: | 2013-11-14 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.524 Å) | | Cite: | Structural basis for promutagenicity of 8-halogenated Guanine.
J.Biol.Chem., 289, 2014
|
|
3IEZ
 
 | | Crystal structure of the RasGAP C-terminal (RGC) domain of IQGAP2 | | Descriptor: | Ras GTPase-activating-like protein IQGAP2, UNKNOWN ATOM OR ION | | Authors: | Nedyalkova, L, Tempel, W, Tong, Y, Zhong, N, Crombet, L, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-23 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the RasGAP C-terminal (RGC) domain
of IQGAP2
To be Published
|
|
7EH8
 
 | | Cryo-EM structure of the hexameric state of C-phycocyanin from Thermoleptolyngbya sp. O-77 | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHYCOCYANOBILIN | | Authors: | Minato, T, Teramoto, T, Adachi, N, Hung, N.K, Yamada, K, Kawasaki, M, Akutsu, M, Moriya, T, Senda, T, Ogo, S, Kakuta, Y, Yoon, K.S. | | Deposit date: | 2021-03-28 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Non-conventional octameric structure of C-phycocyanin.
Commun Biol, 4, 2021
|
|
4NLD
 
 | | Crystal structure of the hepatitis C virus NS5B RNA-dependent RNA polymerase complex with BMS-791325 also known as (1aR,12bS)-8-cyclohexyl-n-(dimethylsulfamoyl)-11-methoxy-1a-{[(1R,5S)-3-methyl-3,8-diazabicyclo[3.2.1]oct-8-yl]carbonyl}-1,1a,2,12b-tetrahydrocyclopropa[d]indolo[2,1-a][2]benzazepine-5-carboxamide and 2-(4-fluorophenyl)-n-methyl-6-[(methylsulfonyl)amino]-5-(propan-2-yloxy)-1-benzofuran-3-carboxamide | | Descriptor: | (1aR,12bS)-8-cyclohexyl-N-(dimethylsulfamoyl)-11-methoxy-1a-{[(1R,5S)-3-methyl-3,8-diazabicyclo[3.2.1]oct-8-yl]carbonyl}-1,1a,2,12b-tetrahydrocyclopropa[d]indolo[2,1-a][2]benzazepine-5-carboxamide, 2-(4-fluorophenyl)-N-methyl-6-[(methylsulfonyl)amino]-5-(propan-2-yloxy)-1-benzofuran-3-carboxamide, RNA-directed RNA polymerase, ... | | Authors: | Sheriff, S. | | Deposit date: | 2013-11-14 | | Release date: | 2014-03-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Discovery and Preclinical Characterization of the Cyclopropylindolobenzazepine BMS-791325, A Potent Allosteric Inhibitor of the Hepatitis C Virus NS5B Polymerase.
J.Med.Chem., 57, 2014
|
|
7EH7
 
 | | Cryo-EM structure of the octameric state of C-phycocyanin from Thermoleptolyngbya sp. O-77 | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHYCOCYANOBILIN | | Authors: | Minato, T, Teramoto, T, Adachi, N, Hung, N.K, Yamada, K, Kawasaki, M, Akutsu, M, Moriya, T, Senda, T, Ogo, S, Kakuta, Y, Yoon, K.S. | | Deposit date: | 2021-03-28 | | Release date: | 2021-11-17 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Non-conventional octameric structure of C-phycocyanin.
Commun Biol, 4, 2021
|
|
8AHM
 
 | | Crystal structure of tubulin in complex with C(13)/C(13')-Bis-Desmethyl-Disorazole Z | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Oliva, M.A, Diaz, J.F, Altmann, K.H. | | Deposit date: | 2022-07-22 | | Release date: | 2022-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Synthesis and Biological Evaluation of C(13)/C(13')-Bis(desmethyl)disorazole Z.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
1XXE
 
 | | RDC refined solution structure of the AaLpxC/TU-514 complex | | Descriptor: | 1,5-ANHYDRO-2-C-(CARBOXYMETHYL-N-HYDROXYAMIDE)-2-DEOXY-3-O-MYRISTOYL-D-GLUCITOL, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Coggins, B.E, McClerren, A.L, Jiang, L, Li, X, Rudolph, J, Hindsgaul, O, Raetz, C.R.H, Zhou, P. | | Deposit date: | 2004-11-04 | | Release date: | 2004-11-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined Solution Structure of the LpxC-TU-514 Complex and pK(a) Analysis of an Active Site Histidine: Insights into the Mechanism and Inhibitor Design
Biochemistry, 44, 2005
|
|
6BBX
 
 | | Crystal structure of TnmS3 in complex with TNM C | | Descriptor: | Glyoxalase/bleomycin resisance protein/dioxygenase, methyl (2R,3R)-2,3-dihydroxy-3-[(1aS,11S,11aR,14Z,18R)-3,7,8,18-tetrahydroxy-4,9-dioxo-4,9,10,11-tetrahydro-11aH-11,1a-hept[3]ene[1,5]diynonaphtho[2,3-h]oxireno[c]quinolin-11a-yl]butanoate | | Authors: | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-10-19 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Resistance to Enediyne Antitumor Antibiotics by Sequestration.
Cell Chem Biol, 25, 2018
|
|
6QNK
 
 | | Antibody FAB fragment targeting Gi protein heterotrimer | | Descriptor: | 1,2-ETHANEDIOL, D-MALATE, FAB heavy chain, ... | | Authors: | Tsai, C.-J, Muehle, J, Pamula, F, Dawson, R.J.P, Maeda, S, Deupi, X, Schertler, G.F.X. | | Deposit date: | 2019-02-11 | | Release date: | 2019-07-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cryo-EM structure of the rhodopsin-G alpha i-beta gamma complex reveals binding of the rhodopsin C-terminal tail to the G beta subunit.
Elife, 8, 2019
|
|
4OBW
 
 | | crystal structure of yeast Coq5 in the SAM bound form | | Descriptor: | 2-methoxy-6-polyprenyl-1,4-benzoquinol methylase, mitochondrial, S-ADENOSYLMETHIONINE, ... | | Authors: | Dai, Y.N, Zhou, K, Cao, D.D, Jiang, Y.L, Meng, F, Chi, C.B, Ren, Y.M, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2014-01-07 | | Release date: | 2014-08-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures and catalytic mechanism of the C-methyltransferase Coq5 provide insights into a key step of the yeast coenzyme Q synthesis pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
