8G1A
 
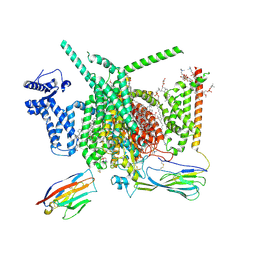 | | Cryo-EM structure of Nav1.7 with CBD | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2023-02-01 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cannabidiol inhibits Na v channels through two distinct binding sites.
Nat Commun, 14, 2023
|
|
6WRU
 
 | |
5MKY
 
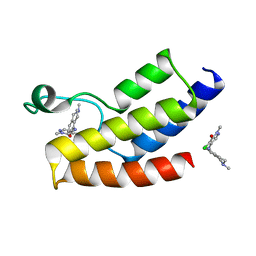 | | BROMODOMAIN OF HUMAN BRD9 WITH 4-chloro-2-methyl-5-((2-methyl-1,2,3,4-tetrahydroisoquinolin-5-yl)amino)pyridazin-3(2H)-one | | Descriptor: | 4-chloranyl-2-methyl-5-[(2-methyl-3,4-dihydro-1~{H}-isoquinolin-5-yl)amino]pyridazin-3-one, Bromodomain-containing protein 9 | | Authors: | Chung, C.-W. | | Deposit date: | 2016-12-05 | | Release date: | 2017-12-20 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Discovery of a Potent, Cell Penetrant, and Selective p300/CBP-Associated Factor (PCAF)/General Control Nonderepressible 5 (GCN5) Bromodomain Chemical Probe.
J. Med. Chem., 60, 2017
|
|
8BVD
 
 | | FimH lectin domain in complex with mannose C-linked to quinoline | | Descriptor: | (2R,3S,4R,5S,6R)-2-(hydroxymethyl)-6-[(E)-3-quinolin-6-ylprop-2-enyl]oxane-3,4,5-triol, Type 1 fimbrin D-mannose specific adhesin | | Authors: | Bouckaert, J, Bridot, C. | | Deposit date: | 2022-12-03 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-08 | | Method: | X-RAY DIFFRACTION (2.995 Å) | | Cite: | Insightful Improvement in the Design of Potent Uropathogenic E. coli FimH Antagonists.
Pharmaceutics, 15, 2023
|
|
5A15
 
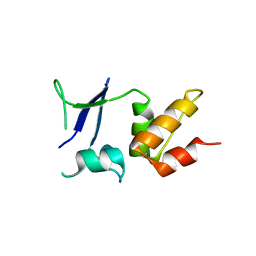 | | Crystal structure of the BTB domain of human KCTD16 | | Descriptor: | BTB/POZ DOMAIN-CONTAINING PROTEIN KCTD16 | | Authors: | Pinkas, D.M, Sanvitale, C.E, Solcan, N, Goubin, S, Canning, P, Dixon Clarke, S.E, Talon, R, Wiggers, H.J, Fitzpatrick, F, Tallant, C, Kopec, J, Chalk, R, Doutch, J, Krojer, T, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2015-04-28 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural complexity in the KCTD family of Cullin3-dependent E3 ubiquitin ligases.
Biochem. J., 474, 2017
|
|
5LKT
 
 | | Crystal structure of the p300 acetyltransferase catalytic core with butyryl-coenzyme A. | | Descriptor: | Butyryl Coenzyme A, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Kaczmarska, Z, Ortega, E, Marquez, J.A, Panne, D. | | Deposit date: | 2016-07-24 | | Release date: | 2016-11-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of p300 in complex with acyl-CoA variants.
Nat. Chem. Biol., 13, 2017
|
|
8EBK
 
 | |
4AQ7
 
 | | Ternary complex of E. coli leucyl-tRNA synthetase, tRNA(leu) and leucyl-adenylate analogue in the aminoacylation conformation | | Descriptor: | E. COLI TRNALEU UAA ISOACCEPTOR, LEUCINE, LEUCINE--TRNA LIGASE, ... | | Authors: | Palencia, A, Crepin, T, Vu, M.T, Lincecum Jr, T.L, Martinis, S.A, Cusack, S. | | Deposit date: | 2012-04-13 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Dynamics of the Aminoacylation and Proofreading Functional Cycle of Bacterial Leucyl-tRNA Synthetase
Nat.Struct.Mol.Biol., 19, 2012
|
|
8A35
 
 | | NaK C-DI mutant with Rb+ and Na+ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Potassium channel protein, RUBIDIUM ION, ... | | Authors: | Minniberger, S, Plested, A.J.R. | | Deposit date: | 2022-06-07 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Asymmetry and Ion Selectivity Properties of Bacterial Channel NaK Mutants Derived from Ionotropic Glutamate Receptors.
J.Mol.Biol., 435, 2023
|
|
8P6O
 
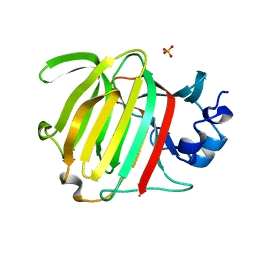 | |
8PC8
 
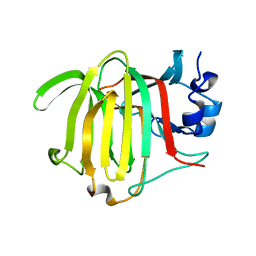 | | Crystal structure of Paradendryphiella salina PL7C alginate lyase soaked with hexamannuronic acid | | Descriptor: | Alginate lyase, beta-D-mannopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-alpha-D-mannopyranuronic acid | | Authors: | Wilknes, C. | | Deposit date: | 2023-06-10 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystal structure of Paradendryphiella salina PL7C alginate lyase soaked with hexamannuronic acid
To Be Published
|
|
8PC3
 
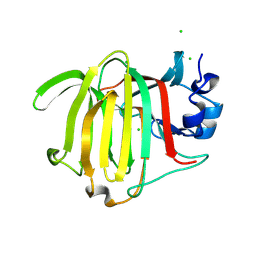 | |
4TPL
 
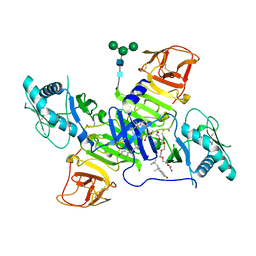 | | West Nile Virus Non-structural protein 1 (NS1) Form 1 crystal | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, OXTOXYNOL-10, ... | | Authors: | Akey, D.L, Smith, J.L. | | Deposit date: | 2014-06-08 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Use of massively multiple merged data for low-resolution S-SAD phasing and refinement of flavivirus NS1.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5LRB
 
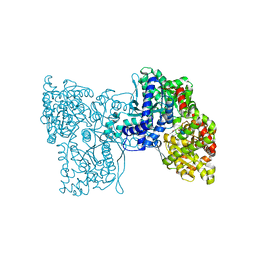 | | Plastidial phosphorylase from Barley in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-1,4 glucan phosphorylase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Cuesta-Seijo, J.A, Ruzanski, C, Kruzewicz, K, Palcic, M.M. | | Deposit date: | 2016-08-18 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Functional and structural characterization of plastidic starch phosphorylase during barley endosperm development.
PLoS ONE, 12, 2017
|
|
4WQ9
 
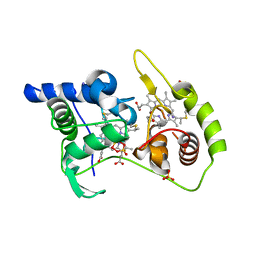 | | Thiosulfate dehydrogenase (TsdA) from Allochromatium vinosum - dithionite soak | | Descriptor: | 1,2-ETHANEDIOL, HEME C, HYDROSULFURIC ACID, ... | | Authors: | Brito, J.A, Denkmann, K, Pereira, I.A.C, Dahl, C, Archer, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-18 | | Last modified: | 2021-09-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Thiosulfate Dehydrogenase (TsdA) from Allochromatium vinosum: STRUCTURAL AND FUNCTIONAL INSIGHTS INTO THIOSULFATE OXIDATION.
J.Biol.Chem., 290, 2015
|
|
8BRE
 
 | |
6FJX
 
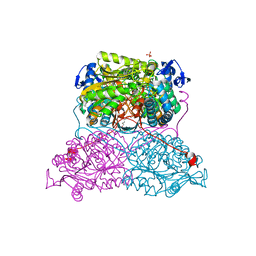 | | Structure and function of aldehyde dehydrogenase from Thermus thermophilus: An enzyme with an evolutionarily-distinct C-terminal arm (Native protein) | | Descriptor: | Aldehyde dehydrogenase, SULFATE ION, TRIETHYLENE GLYCOL, ... | | Authors: | Hayes, K.A, Noor, M.R, Djeghader, A, Soulimane, T. | | Deposit date: | 2018-01-23 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The quaternary structure of Thermus thermophilus aldehyde dehydrogenase is stabilized by an evolutionary distinct C-terminal arm extension.
Sci Rep, 8, 2018
|
|
6WF7
 
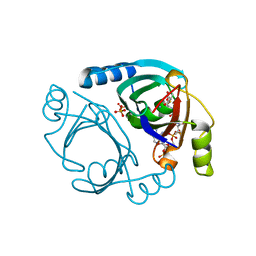 | | Methylmalonyl-CoA epimerase in complex with methylmalonyl-CoA and NH4+ | | Descriptor: | (S)-Methylmalonyl-Coenzyme A, AMMONIUM ION, METHYLMALONYL-COENZYME A, ... | | Authors: | Stunkard, L.M, Benjamin, A.B, Bower, J.B, Huth, T.J, Lohman, J.R. | | Deposit date: | 2020-04-03 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Substrate Enolate Intermediate and Mimic Captured in the Active Site of Streptomyces coelicolor Methylmalonyl-CoA Epimerase.
Chembiochem, 23, 2022
|
|
5KU3
 
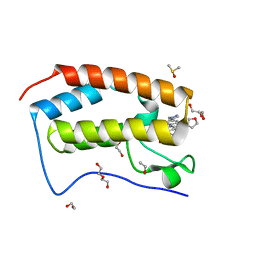 | | BRD4 bromodomain in complex with Cpd59 ((S)-1-(3-((2-fluoro-4-(1-methyl-1H-pyrazol-4-yl)phenyl)amino)-1-(tetrahydrofuran-3-yl)-6,7-dihydro-1H-pyrazolo[4,3-c]pyridin-5(4H)-yl)ethanone) | | Descriptor: | 1,2-ETHANEDIOL, 1-[3-[[2-fluoranyl-4-(1-methylpyrazol-4-yl)phenyl]amino]-1-[(3~{S})-oxolan-3-yl]-6,7-dihydro-4~{H}-pyrazolo[4,3-c]pyridin-5-yl]ethanone, Bromodomain-containing protein 4, ... | | Authors: | Murray, J.M, Huang, W. | | Deposit date: | 2016-07-12 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Discovery of a Potent and Selective in Vivo Probe (GNE-272) for the Bromodomains of CBP/EP300.
J. Med. Chem., 59, 2016
|
|
6WVH
 
 | | Human VKOR with Brodifacoum | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Brodifacoum, Vitamin K epoxide reductase, ... | | Authors: | Liu, S, Sukumar, N, Li, W. | | Deposit date: | 2020-05-06 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation.
Science, 371, 2021
|
|
3QHN
 
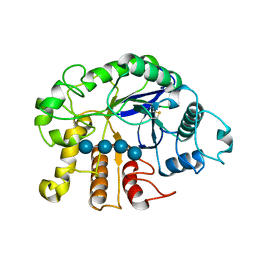 | | Crystal analysis of the complex structure, E201A-cellotetraose, of endocellulase from pyrococcus horikoshii | | Descriptor: | 458aa long hypothetical endo-1,4-beta-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Kim, H.-W, Ishikawa, K. | | Deposit date: | 2011-01-26 | | Release date: | 2012-02-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Functional analysis of hyperthermophilic endocellulase from Pyrococcus horikoshii by crystallographic snapshots
Biochem.J., 437, 2011
|
|
5KJ0
 
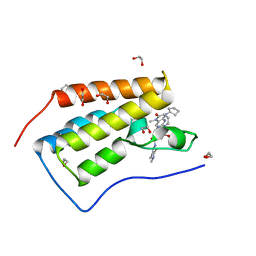 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH DB-1-264-2 | | Descriptor: | 1,2-ETHANEDIOL, 4-[[(7~{R})-8-cyclopentyl-7-ethyl-5-methyl-6-oxidanylidene-7~{H}-pteridin-2-yl]-methyl-amino]-3-methoxy-~{N}-(1-methylpiperidin-4-yl)benzamide, Bromodomain-containing protein 4 | | Authors: | Zhu, J.-Y, Ember, S.W, Schonbrunn, E. | | Deposit date: | 2016-06-17 | | Release date: | 2017-08-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Assessment of Bromodomain Target Engagement by a Series of BI2536 Analogues with Miniaturized BET-BRET.
ChemMedChem, 11, 2016
|
|
7JHD
 
 | | Estrogen Receptor Alpha Ligand Binding Domain Y537S in Complex with TTC-352 and GRIP Peptide | | Descriptor: | 3-(4-fluorophenyl)-2-(4-hydroxyphenoxy)-1-benzothiophene-6-ol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Fanning, S.W, Abderraman, B, Maximov, P.Y, Jordan, V.C, Greene, G.L. | | Deposit date: | 2020-07-20 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Rapid Induction of the Unfolded Protein Response and Apoptosis by Estrogen Mimic TTC-352 for the Treatment of Endocrine-Resistant Breast Cancer.
Mol.Cancer Ther., 20, 2021
|
|
4WOV
 
 | | CRYSTAL STRUCTURE OF TYROSINE KINASE 2 JH2 (PSEUDO KINASE DOMAIN) COMPLEXED WITH BMS-066 AKA 2-METHOXY-N-({6-[3-METHYL-7-(METHYLAMINO)-3,5,8,10-TETRAAZATRICYCLO[7.3.0.0, 6]DODECA-1(9),2(6),4,7,11-PENTAEN-11-YL]PYRIDIN-2-YL}METHY L)ACETAMIDE | | Descriptor: | 2-methoxy-N-({6-[1-methyl-4-(methylamino)-1,6-dihydroimidazo[4,5-d]pyrrolo[2,3-b]pyridin-7-yl]pyridin-2-yl}methyl)acetamide, Non-receptor tyrosine-protein kinase TYK2, SULFATE ION | | Authors: | Muckelbauer, J.K. | | Deposit date: | 2014-10-16 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Tyrosine Kinase 2-mediated Signal Transduction in T Lymphocytes Is Blocked by Pharmacological Stabilization of Its Pseudokinase Domain.
J.Biol.Chem., 290, 2015
|
|
5J03
 
 | |
