3UU4
 
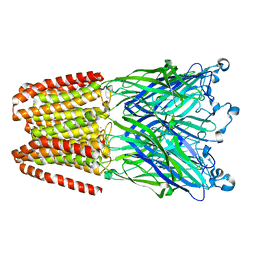 | | The GLIC pentameric Ligand-Gated Ion Channel Loop2-21' mutant reduced in the crystal in a locally-closed conformation (LC1 subtype) | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Glr4197 protein | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-11-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3UUM
 
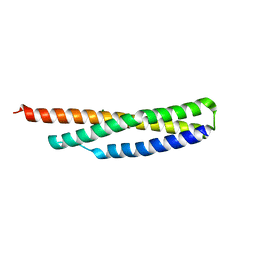 | |
4O6Y
 
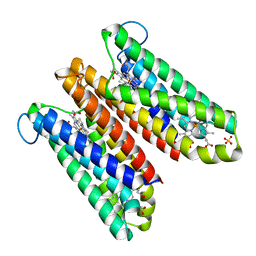 | | Crystal Structure of Cytochrome b561 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Probable transmembrane ascorbate ferrireductase 2, SULFATE ION | | Authors: | Lu, P, Ma, D, Yan, C, Gong, X, Du, M, Shi, Y. | | Deposit date: | 2013-12-24 | | Release date: | 2014-02-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and mechanism of a eukaryotic transmembrane ascorbate-dependent oxidoreductase
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3UWW
 
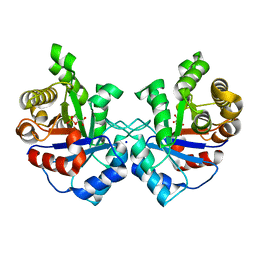 | | Crystal structure of Staphylococcus Aureus triosephosphate isomerase complexed with 3-phosphoglyceric acid | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 3-PHOSPHOGLYCERIC ACID, SODIUM ION, ... | | Authors: | Mukherjee, S, Roychowdhury, A, Dutta, D, Das, A.K. | | Deposit date: | 2011-12-03 | | Release date: | 2012-10-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of triosephosphate isomerase from methicillin resistant Staphylococcus aureus MRSA252 provide structural insights into novel modes of ligand binding and unique conformations of catalytic loop
Biochimie, 94, 2012
|
|
4OJ1
 
 | |
3UV3
 
 | | Ec_IspH in complex with but-2-ynyl diphosphate (1086) | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER, but-2-yn-1-yl trihydrogen diphosphate | | Authors: | Span, I, Wang, K, Wang, W, Zhang, Y, Bacher, A, Eisenreich, W, Schulz, C, Oldfield, E, Groll, M. | | Deposit date: | 2011-11-29 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of acetylene hydratase activity of the iron-sulphur protein IspH.
Nat Commun, 3, 2012
|
|
4O7B
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with SB-284847-BT | | Descriptor: | 1,2-ETHANEDIOL, 2-(2,3-dimethylphenoxy)-4-[4-(4-fluorophenyl)-1-(piperidin-4-yl)-1H-imidazol-5-yl]pyrimidine, Bromodomain-containing protein 4 | | Authors: | Ember, S.W, Zhu, J.-Y, Watts, C, Schonbrunn, E. | | Deposit date: | 2013-12-24 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
4O7J
 
 | |
3UVN
 
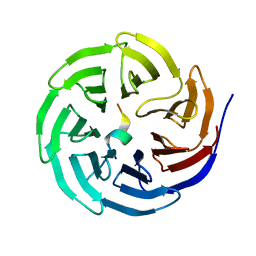 | | Crystal structure of WDR5 in complex with the WDR5-interacting motif of SET1A | | Descriptor: | Histone-lysine N-methyltransferase SETD1A, WD repeat-containing protein 5 | | Authors: | Zhang, P, Lee, H, Brunzelle, J.S, Couture, J.-F. | | Deposit date: | 2011-11-30 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | The plasticity of WDR5 peptide-binding cleft enables the binding of the SET1 family of histone methyltransferases.
Nucleic Acids Res., 40, 2012
|
|
3UXY
 
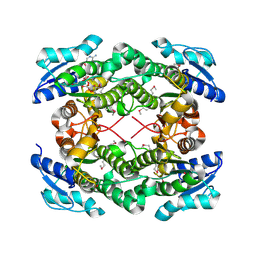 | | The crystal structure of short chain dehydrogenase from Rhodobacter sphaeroides | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Short-chain dehydrogenase/reductase SDR | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-05 | | Release date: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | The crystal structure of short chain dehydrogenase from Rhodobacter sphaeroides
To be Published
|
|
3UWB
 
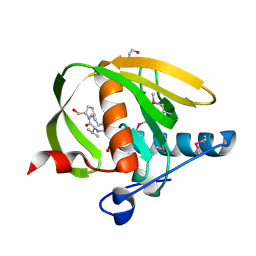 | | Crystal structure of a probable peptide deformylase from strucynechococcus phage S-SSM7 in complex with actinonin | | Descriptor: | 1,2-ETHANEDIOL, ACTINONIN, CHLORIDE ION, ... | | Authors: | Lorimer, D, Abendroth, J, Edwards, T.E, Burgin, A, Segall, A, Rohwer, F. | | Deposit date: | 2011-12-01 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and function of a cyanophage-encoded peptide deformylase.
ISME J, 7, 2013
|
|
4OJN
 
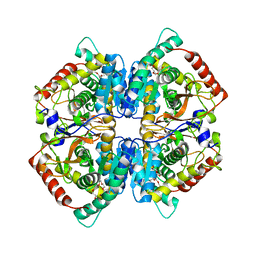 | | Crystal structure of human muscle L-lactate dehydrogenase | | Descriptor: | GLYCEROL, L-lactate dehydrogenase A chain, PENTAETHYLENE GLYCOL | | Authors: | Kolappan, S, Craig, L. | | Deposit date: | 2014-01-21 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of lactate dehydrogenase A (LDHA) in apo, ternary and inhibitor-bound forms.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3UYR
 
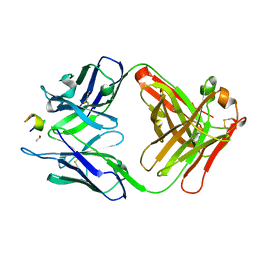 | | Structure of a monoclonal antibody complexed with its MHC-I antigen | | Descriptor: | 1,2-ETHANEDIOL, H-2 class I histocompatibility antigen, L-D alpha chain, ... | | Authors: | Margulies, D.H, Mage, M.G, Wang, R, Natarajan, K. | | Deposit date: | 2011-12-06 | | Release date: | 2012-07-25 | | Last modified: | 2012-08-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Peptide-receptive transition state of MHC class I molecules: insight from structure and molecular dynamics.
J.Immunol., 189, 2012
|
|
4O89
 
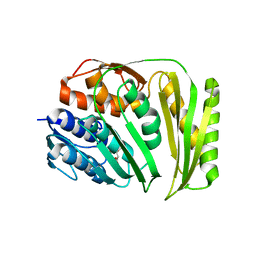 | | Crystal structure of RtcA, the RNA 3'-terminal phosphate cyclase from Pyrococcus horikoshii. | | Descriptor: | CITRIC ACID, RNA 3'-terminal phosphate cyclase | | Authors: | Desai, K.K, Bingman, C.A, Phillips Jr, G.N, Raines, R.T. | | Deposit date: | 2013-12-26 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of RNA 3'-phosphate cyclase bound to substrate RNA.
Rna, 20, 2014
|
|
2R94
 
 | | Crystal Structure of KD(P)GA from T.tenax | | Descriptor: | 2-Keto-3-deoxy-(6-phospho-)gluconate aldolase, PYRUVIC ACID | | Authors: | Pauluhn, A, Pohl, E. | | Deposit date: | 2007-09-12 | | Release date: | 2008-03-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and stereochemical studies of KD(P)G aldolase from Thermoproteus tenax.
Proteins, 72, 2008
|
|
3UWL
 
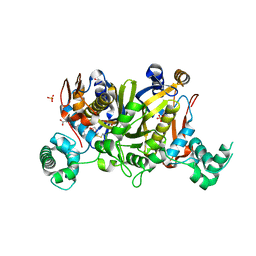 | | Crystal structure of Enteroccocus faecalis thymidylate synthase (EfTS) in complex with 5-formyl tetrahydrofolate | | Descriptor: | 1,2-ETHANEDIOL, N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, SULFATE ION, ... | | Authors: | Pozzi, C, Catalano, A, Cortesi, D, Luciani, R, Ferrari, S, Fritz, T, Costi, M.P, Mangani, S. | | Deposit date: | 2011-12-02 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The structure of Enterococcus faecalis thymidylate synthase provides clues about folate bacterial metabolism.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4O8W
 
 | | Crystal Structure of the GerD spore germination protein | | Descriptor: | Spore germination protein | | Authors: | Li, Y, Jin, K, Ghosh, S, Devarakonda, P, Carlson, K, Davis, A, Stewart, K, Cammett, E, Rossi, P.P, Setlow, B, Lu, M, Setlow, P, Hao, B. | | Deposit date: | 2013-12-30 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Structural and Functional Analysis of the GerD Spore Germination Protein of Bacillus Species.
J.Mol.Biol., 426, 2014
|
|
3UXW
 
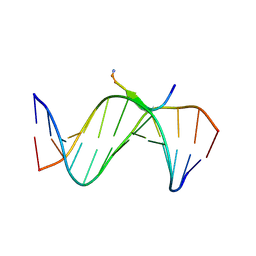 | | Crystal Structures of an A-T-hook/DNA complex | | Descriptor: | A-T hook peptide, dodecamer DNA | | Authors: | Fonfria-Subiros, E, Acosta-Reyes, F.J, Saperas, N, Pous, J, Subirana, J.A, Campos, J.L. | | Deposit date: | 2011-12-05 | | Release date: | 2012-05-23 | | Last modified: | 2013-03-27 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of a complex of DNA with one AT-hook of HMGA1.
Plos One, 7, 2012
|
|
3V05
 
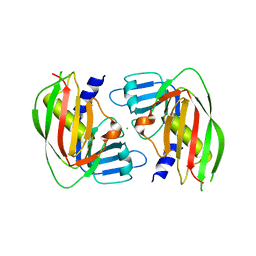 | | 2.4 Angstrom Crystal Structure of Superantigen-like Protein from Staphylococcus aureus. | | Descriptor: | CHLORIDE ION, Superantigen-like Protein | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Filippova, E.V, Dubrovska, I, Winsor, J, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-07 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4 Angstrom Crystal Structure of Superantigen-like Protein from Staphylococcus aureus.
TO BE PUBLISHED
|
|
4O9E
 
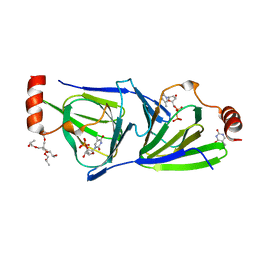 | | Crystal structure of QdtA, a sugar 3,4-ketoisemerase from Thermoanaerobacterium thermosaccharolyticum in complex with TDP | | Descriptor: | (2S)-1-[3-[(2S)-2-oxidanylpropoxy]-2-[[(2S)-2-oxidanylpropoxy]methyl]-2-[[(2R)-2-oxidanylpropoxy]methyl]propoxy]propan-2-ol, QdtA, THYMIDINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Holden, H.M. | | Deposit date: | 2014-01-02 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The molecular architecture of QdtA, a sugar 3,4-ketoisomerase from Thermoanaerobacterium thermosaccharolyticum.
Protein Sci., 23, 2014
|
|
3UY9
 
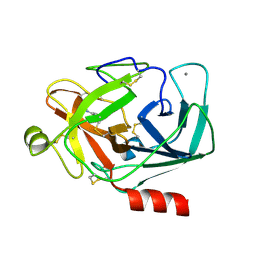 | | Bovine trypsin variant X(tripleGlu217Phe227) in complex with small molecule inhibitor | | Descriptor: | BENZAMIDINE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Tziridis, A, Neumann, P, Kolenko, P, Stubbs, M.T. | | Deposit date: | 2011-12-06 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Correlating structure and ligand affinity in drug discovery: a cautionary tale involving second shell residues.
Biol.Chem., 395, 2014
|
|
4OKT
 
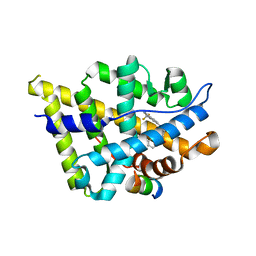 | | Crystal structure of W741L-AR-LBD bound with co-regulator peptide | | Descriptor: | Androgen receptor, R-BICALUTAMIDE, co-regulator peptide | | Authors: | Liu, J.S, Hsu, C.L, Wu, W.G. | | Deposit date: | 2014-01-22 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
4O9P
 
 | | Crystal structure of Thermus thermophilis transhydrogeanse domain II dimer SeMet derivative | | Descriptor: | NAD(P) transhydrogenase subunit alpha 2, NAD(P) transhydrogenase subunit beta | | Authors: | Leung, J.H, Yamaguchi, M, Moeller, A, Schurig-Briccio, L.A, Gennis, R.B, Potter, C.S, Carragher, B, Stout, C.D. | | Deposit date: | 2014-01-02 | | Release date: | 2014-06-11 | | Last modified: | 2015-01-28 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structural biology. Division of labor in transhydrogenase by alternating proton translocation and hydride transfer.
Science, 347, 2015
|
|
4OL0
 
 | | Crystal structure of transportin-SR2, a karyopherin involved in human disease, in complex with Ran | | Descriptor: | GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tsirkone, V.G, Strelkov, S.V. | | Deposit date: | 2014-01-23 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of transportin SR2, a karyopherin involved in human disease, in complex with Ran.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
3V0D
 
 | | Crystal structure of Ciona intestinalis voltage sensor-containing phosphatase (Ci-VSP), residues 241-576(C363S) | | Descriptor: | PHOSPHATE ION, Voltage-sensor containing phosphatase | | Authors: | Liu, L, Kohout, S.C, Xu, Q, Muller, S, Kimberlin, C, Isacoff, E.Y, Minor, D.L. | | Deposit date: | 2011-12-07 | | Release date: | 2012-05-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | A glutamate switch controls voltage-sensitive phosphatase function.
Nat.Struct.Mol.Biol., 19, 2012
|
|
