8FTM
 
 | | Setx-ssRNA-ADP-SO4 complex | | Descriptor: | 5'-3' RNA helicase-like protein, ADENOSINE-5'-DIPHOSPHATE, RNA (5'-R(*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3'), ... | | Authors: | Williams, R.S, Appel, C.D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Sen1 architecture: RNA-DNA hybrid resolution, autoregulation, and insights into SETX inactivation in AOA2.
Mol.Cell, 83, 2023
|
|
6N8H
 
 | |
3BT7
 
 | |
4X5O
 
 | | Human histidine tRNA synthetase | | Descriptor: | Histidine--tRNA ligase, cytoplasmic | | Authors: | Kim, Y.K, Jeon, Y.H. | | Deposit date: | 2014-12-05 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural characteristics of human histidyl-tRNA synthetase
Biodesign, 2, 2015
|
|
8WUU
 
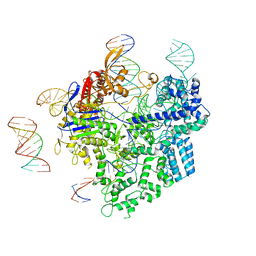 | | SpCas9-pegRNA-target DNA complex (pre-initiation) | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (34-MER), DNA (5'-D(*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), ... | | Authors: | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2023-10-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
3TUP
 
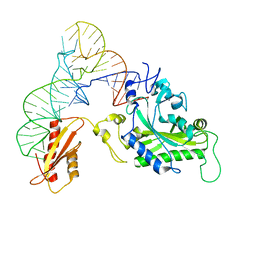 | | Crystal structure of human mitochondrial PheRS complexed with tRNAPhe in the active open state | | Descriptor: | Phenylalanyl-tRNA synthetase, mitochondrial, Thermus thermophilus tRNAPhe | | Authors: | Safro, M, Klipcan, L, Moor, N, Finarov, I, Kessler, N, Sukhanova, M. | | Deposit date: | 2011-09-17 | | Release date: | 2011-11-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal Structure of Human Mitochondrial PheRS Complexed with tRNA(Phe) in the Active "Open" State.
J.Mol.Biol., 415, 2012
|
|
2LBL
 
 | |
7LO9
 
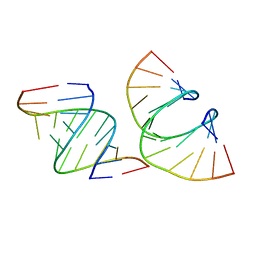 | | RNA dodecamer containing a GNA A residue | | Descriptor: | Chains: A,B,C,D | | Authors: | Harp, J.M, Wawrzak, Z, Egli, M. | | Deposit date: | 2021-02-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Overcoming GNA/RNA base-pairing limitations using isonucleotides improves the pharmacodynamic activity of ESC+ GalNAc-siRNAs.
Nucleic Acids Res., 49, 2021
|
|
8IEW
 
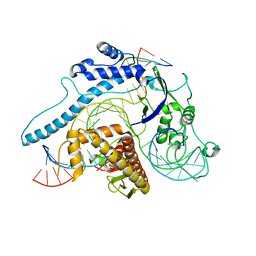 | | Cas005-crRNA-DNA complex | | Descriptor: | Cas005, DNA (55-MER), RNA (38-MER) | | Authors: | Zhang, X, Duan, Z.Q, Zhu, J.K. | | Deposit date: | 2023-02-16 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of Cas005-crRNA-DNA complex at 3.1 Angstroms resolution
To Be Published
|
|
5DO5
 
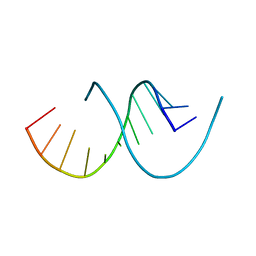 | |
3IYR
 
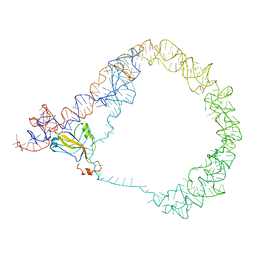 | | tmRNA-SmpB: a journey to the center of the bacterial ribosome | | Descriptor: | SsrA-binding protein, tmRNA | | Authors: | Weis, F, Bron, P, Giudice, E, Rolland, J.P, Thomas, D, Felden, B, Gillet, R. | | Deposit date: | 2010-04-16 | | Release date: | 2010-10-20 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | tmRNA-SmpB: a journey to the centre of the bacterial ribosome.
Embo J., 29, 2010
|
|
6E9E
 
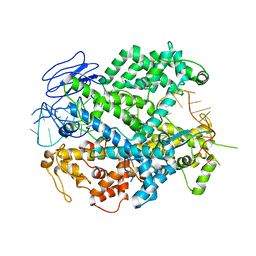 | | EsCas13d-crRNA binary complex | | Descriptor: | EsCas13d, MAGNESIUM ION, crRNA (52-MER) | | Authors: | Zhang, C, Lyumkis, D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Basis for the RNA-Guided Ribonuclease Activity of CRISPR-Cas13d.
Cell, 175, 2018
|
|
3IYQ
 
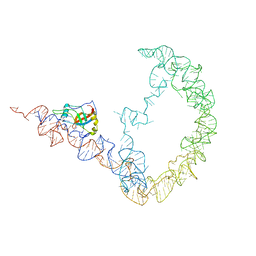 | | tmRNA-SmpB: a journey to the center of the bacterial ribosome | | Descriptor: | SsrA-binding protein, tmRNA | | Authors: | Weis, F, Bron, P, Giudice, E, Rolland, J.P, Thomas, D, Felden, B, Gillet, R. | | Deposit date: | 2010-04-16 | | Release date: | 2010-10-20 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | tmRNA-SmpB: a journey to the centre of the bacterial ribosome.
Embo J., 29, 2010
|
|
4PNO
 
 | |
2NNW
 
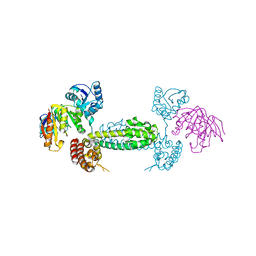 | | Alternative conformations of Nop56/58-fibrillarin complex and implication for induced-fit assenly of box C/D RNPs | | Descriptor: | Fibrillarin-like rRNA/tRNA 2'-O-methyltransferase, NOP5/NOP56 related protein | | Authors: | Oruganti, S, Zhang, Y, Terns, R, Terns, M.P, Li, H. | | Deposit date: | 2006-10-24 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Alternative Conformations of the Archaeal Nop56/58-Fibrillarin Complex Imply Flexibility in Box C/D RNPs.
J.Mol.Biol., 371, 2007
|
|
5VAJ
 
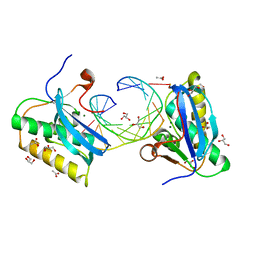 | | BhRNase H - amide-RNA/DNA complex | | Descriptor: | ACETATE ION, DNA (5'-D(*GP*AP*AP*TP*CP*AP*GP*GP*TP*GP*TP*C)-3'), GLYCEROL, ... | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2017-03-27 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Amide linkages mimic phosphates in RNA interactions with proteins and are well tolerated in the guide strand of short interfering RNAs.
Nucleic Acids Res., 45, 2017
|
|
3W1H
 
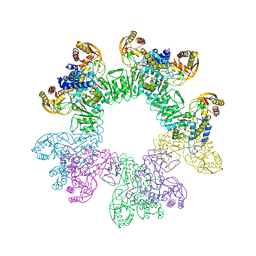 | |
6Y6M
 
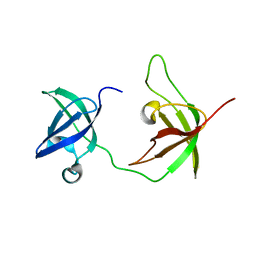 | |
5AMQ
 
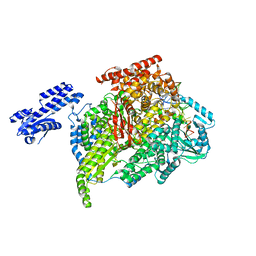 | |
5AMR
 
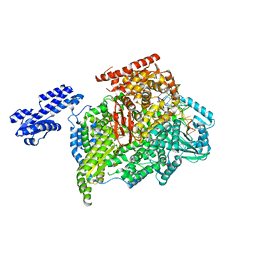 | |
1Z5F
 
 | | Solution Structure of the Cytotoxic RC-RNase 3 with a Pyroglutamate Residue at the N-terminus | | Descriptor: | RC-RNase 3 | | Authors: | Lou, Y.C, Huang, Y.C, Pan, Y.R, Chen, C, Liao, Y.D. | | Deposit date: | 2005-03-18 | | Release date: | 2006-02-28 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Roles of N-terminal pyroglutamate in maintaining structural integrity and pKa values of catalytic histidine residues in bullfrog ribonuclease 3
J.Mol.Biol., 355, 2006
|
|
3W1J
 
 | |
3W1I
 
 | |
3CUL
 
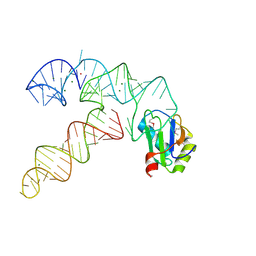 | | Aminoacyl-tRNA synthetase ribozyme | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, RNA (92-MER), ... | | Authors: | Xiao, H, Murakami, H, Suga, H, Ferre-D'Amare, A.R. | | Deposit date: | 2008-04-16 | | Release date: | 2008-06-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of specific tRNA aminoacylation by a small in vitro selected ribozyme.
Nature, 454, 2008
|
|
6Y96
 
 | | solution structure of cold-shock domain 9 of drosophila Upstream of N-Ras (Unr) | | Descriptor: | Upstream of N-ras, isoform A | | Authors: | Sweetapple, L.J, Hollmann, N.M, Simon, B, Hennig, J. | | Deposit date: | 2020-03-06 | | Release date: | 2020-07-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Pseudo-RNA-Binding Domains Mediate RNA Structure Specificity in Upstream of N-Ras.
Cell Rep, 32, 2020
|
|
