6NLZ
 
 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with fragment degradation product B9D | | Descriptor: | ATP-dependent dethiobiotin synthetase BioD, SULFATE ION, [(1R,2S)-2-(2-hydroxybenzene-1-carbonyl)cyclopentyl]acetic acid, ... | | Authors: | Thompson, A.P, Polyak, S.W, Wegener, K.L, Bruning, J.B. | | Deposit date: | 2019-01-10 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with fragment degradation product B9D
To Be Published
|
|
6GI3
 
 | |
6W84
 
 | | K2P2.1 (TREK-1), 200 mM K+ | | Descriptor: | CADMIUM ION, DODECANE, N-OCTANE, ... | | Authors: | Lolicato, M, Minor, D.L. | | Deposit date: | 2020-03-20 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | K 2P channel C-type gating involves asymmetric selectivity filter order-disorder transitions.
Sci Adv, 6, 2020
|
|
5EP5
 
 | |
6VV9
 
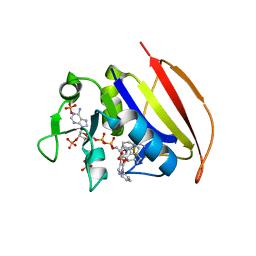 | | Mycobacterium tuberculosis dihydrofolate reductase in complex with JEB300 | | Descriptor: | 5-[4-(1H-indol-3-yl)butoxy]-1-phenyl-1H-pyrazole-4-carboxylic acid, COBALT (II) ION, Dihydrofolate reductase, ... | | Authors: | Ribeiro, J.A, Chavez-Pacheco, S.M, Dias, M.V.B. | | Deposit date: | 2020-02-17 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Using a Fragment-Based Approach to Identify Alternative Chemical Scaffolds Targeting Dihydrofolate Reductase fromMycobacterium tuberculosis.
Acs Infect Dis., 6, 2020
|
|
5UV5
 
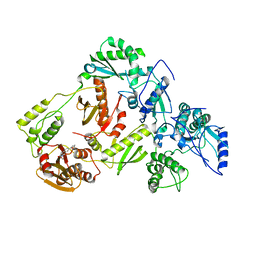 | | Crystal Structure of a 2-Hydroxyisoquinoline-1,3-dione RNase H Active Site Inhibitor with Multiple Binding Modes to HIV Reverse Transcriptase | | Descriptor: | 7-(furan-2-yl)-2-hydroxyisoquinoline-1,3(2H,4H)-dione, MANGANESE (II) ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2017-02-19 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A 2-Hydroxyisoquinoline-1,3-Dione Active-Site RNase H Inhibitor Binds in Multiple Modes to HIV-1 Reverse Transcriptase.
Antimicrob. Agents Chemother., 61, 2017
|
|
5EPJ
 
 | | Crystal Structure of chromodomain of CBX7 in complex with inhibitor UNC3866 | | Descriptor: | Chromobox protein homolog 7, UNKNOWN ATOM OR ION, peptide-like inhibitor UNC3866 | | Authors: | Liu, Y, Tempel, W, Walker, J.R, Stuckey, J.I, Dickson, B.M, James, L.I, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-11 | | Release date: | 2015-12-16 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of chromodomain of CBX7 in complex with inhibitor UNC3866
to be published
|
|
6VVU
 
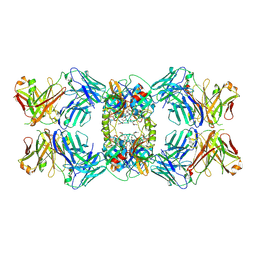 | | Anti-Tryptase fab E104.v1 bound to tryptase | | Descriptor: | CALCIUM ION, Fab E104.v1 heavy chain, Fab E104.v1 light chain, ... | | Authors: | Ultsch, M, Koerber, J.T. | | Deposit date: | 2020-02-18 | | Release date: | 2020-12-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Bivalent antibody pliers inhibit beta-tryptase by an allosteric mechanism dependent on the IgG hinge.
Nat Commun, 11, 2020
|
|
5L1X
 
 | | Structure of the Human Metapneumovirus Fusion Protein in the Postfusion Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mas, V, Melero, J.A, McLellan, J.S. | | Deposit date: | 2016-07-29 | | Release date: | 2016-08-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Engineering, Structure and Immunogenicity of the Human Metapneumovirus F Protein in the Postfusion Conformation.
Plos Pathog., 12, 2016
|
|
6W8A
 
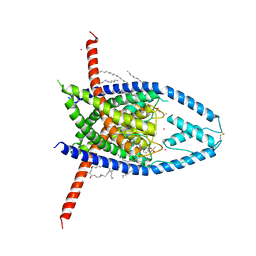 | | K2P2.1 (TREK-1):ML335 complex, 10 mM K+ | | Descriptor: | CADMIUM ION, DODECANE, HEXADECANE, ... | | Authors: | Lolicato, M, Minor, D.L. | | Deposit date: | 2020-03-20 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | K 2P channel C-type gating involves asymmetric selectivity filter order-disorder transitions.
Sci Adv, 6, 2020
|
|
6O04
 
 | | M.tb MenD IntII bound with Inhibitor | | Descriptor: | (1~{R},2~{S},5~{S},6~{S})-2-[(1~{S})-1-[3-[(4-azanylidene-2-methyl-1~{H}-pyrimidin-5-yl)methyl]-4-methyl-5-[2-[oxidanyl (phosphonooxy)phosphoryl]oxyethyl]-1,3-thiazol-3-ium-2-yl]-1,4-bis(oxidanyl)-4-oxidanylidene-butyl]-6-oxidanyl-5-(3-oxid anyl-3-oxidanylidene-prop-1-en-2-yl)oxy-cyclohex-3-ene-1-carboxylic acid, 1,4-dihydroxy-2-naphthoic acid, 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase, ... | | Authors: | Johnston, J.M, Bashiri, G, Bulloch, E.M, Jirgis, E.M.N, Nigon, L.V, Chuang, H, Baker, E.N. | | Deposit date: | 2019-02-15 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Allosteric regulation of menaquinone (vitamin K2) biosynthesis in the human pathogenMycobacterium tuberculosis.
J.Biol.Chem., 295, 2020
|
|
8PMS
 
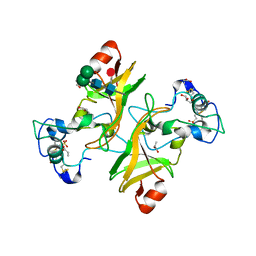 | | NADase from Aspergillus fumigatus with replaced C-terminus from Neurospora crassa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kallio, J.P, Ferrario, E, Stromland, O, Ziegler, M. | | Deposit date: | 2023-06-29 | | Release date: | 2023-11-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Novel Calcium-Binding Motif Stabilizes and Increases the Activity of Aspergillus fumigatus Ecto-NADase.
Biochemistry, 62, 2023
|
|
6XVM
 
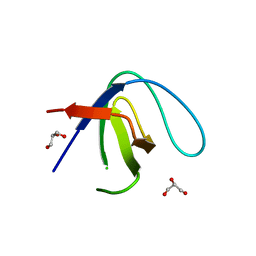 | |
6W8F
 
 | | K2P2.1 (TREK-1):ML335 complex, 0 mM K+ | | Descriptor: | CADMIUM ION, DODECANE, HEXADECANE, ... | | Authors: | Lolicato, M, Minor, D.L. | | Deposit date: | 2020-03-20 | | Release date: | 2021-01-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | K 2P channel C-type gating involves asymmetric selectivity filter order-disorder transitions.
Sci Adv, 6, 2020
|
|
6GJB
 
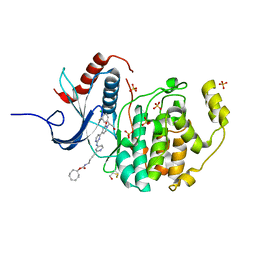 | | Erk2 signalling protein | | Descriptor: | Mitogen-activated protein kinase 1, SULFATE ION, [(1~{R},4~{Z})-cyclooct-4-en-1-yl] ~{N}-[4-[4-[[4-[1-[(1~{S})-1-(4-chloranyl-3-fluoranyl-phenyl)-2-oxidanyl-ethyl]-2-oxidanylidene-pyridin-4-yl]pyrimidin-2-yl]amino]pyridin-2-yl]but-3-ynyl]carbamate | | Authors: | O'Reilly, M. | | Deposit date: | 2018-05-16 | | Release date: | 2019-01-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Quantitation of ERK1/2 inhibitor cellular target occupancies with a reversible slow off-rate probe.
Chem Sci, 9, 2018
|
|
6W8S
 
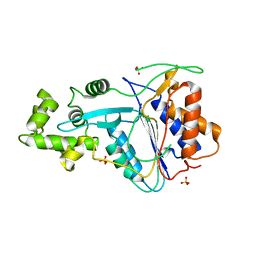 | | Crystal structure of metacaspase 4 from Arabidopsis | | Descriptor: | Metacaspase-4, SULFATE ION | | Authors: | Zhu, P, Yu, X.H, Wang, C, Zhang, Q, Liu, W, McSweeney, S, Shanklin, J, Lam, E, Liu, Q. | | Deposit date: | 2020-03-21 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.484 Å) | | Cite: | Structural basis for Ca2+-dependent activation of a plant metacaspase.
Nat Commun, 11, 2020
|
|
6NN0
 
 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with 2'-deoxycytidine and fragment degradation product B9D | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, ATP-dependent dethiobiotin synthetase BioD, SULFATE ION, ... | | Authors: | Thompson, A.P, Polyak, S.W, Wegener, K.L, Bruning, J.B. | | Deposit date: | 2019-01-14 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.343 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with 2'-deoxycytidine and B9D
To Be Published
|
|
6XPT
 
 | |
6GJM
 
 | | Cyclophilin A complexed with tri-vector ligand 4. | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A, ethyl 2-[[(4-aminophenyl)methyl-propyl-carbamoyl]amino]ethanoate | | Authors: | Georgiou, C, De Simone, A, Walkinshaw, M.D, Michel, J. | | Deposit date: | 2018-05-16 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.354 Å) | | Cite: | A computationally designed binding mode flip leads to a novel class of potent tri-vector cyclophilin inhibitors.
Chem Sci, 10, 2019
|
|
6XQS
 
 | | Room-temperature X-ray Crystal structure of SARS-CoV-2 main protease in complex with Telaprevir | | Descriptor: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, 3C-like proteinase | | Authors: | Kneller, D.W, Kovalevsky, A, Coates, L. | | Deposit date: | 2020-07-10 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Malleability of the SARS-CoV-2 3CL M pro Active-Site Cavity Facilitates Binding of Clinical Antivirals.
Structure, 28, 2020
|
|
5LA0
 
 | | The mechanism by which arabinoxylanases can recognise highly decorated xylans | | Descriptor: | CALCIUM ION, Carbohydrate binding family 6, SULFATE ION, ... | | Authors: | Basle, A, Labourel, A, Cuskin, F, Jackson, A, Crouch, L, Rogowski, A, Gilbert, A. | | Deposit date: | 2016-06-13 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Mechanism by Which Arabinoxylanases Can Recognize Highly Decorated Xylans.
J.Biol.Chem., 291, 2016
|
|
6NNH
 
 | | Structure of Closed state of Dihydrofolate reductase from Mycobacterium tuberculosis in complex with NADPH and cycloguanil | | Descriptor: | 1-(4-chlorophenyl)-6,6-dimethyl-1,6-dihydro-1,3,5-triazine-2,4-diamine, COBALT (II) ION, Dihydrofolate reductase, ... | | Authors: | Giudice, J.H.P, Ribeiro, J.A, Dias, M.V.B. | | Deposit date: | 2019-01-15 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.523 Å) | | Cite: | Crystal structures of the closed form of Mycobacterium tuberculosis dihydrofolate reductase in complex with dihydrofolate and antifolates.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6XHG
 
 | | Far-red absorbing dark state of JSC1_58120g3 with bound biliverdin IXa (BV) | | Descriptor: | 1,2-ETHANEDIOL, 3-[2-[(~{Z})-[5-[(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-pyrrol-2-ylidene]methyl]-5-[(~{Z})-(3-ethyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, JSC1_58120g3 | | Authors: | Moreno, M.V, Rockwell, N.C, Fisher, A.J, Lagarias, J.C. | | Deposit date: | 2020-06-18 | | Release date: | 2020-10-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A far-red cyanobacteriochrome lineage specific for verdins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6W9O
 
 | |
6O4C
 
 | | Structure of ALDH7A1 mutant W175A complexed with NAD | | Descriptor: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde dehydrogenase, CHLORIDE ION, ... | | Authors: | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | Deposit date: | 2019-02-28 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical consequences of pyridoxine-dependent epilepsy mutations that target the aldehyde binding site of aldehyde dehydrogenase ALDH7A1.
Febs J., 287, 2020
|
|
