3UG3
 
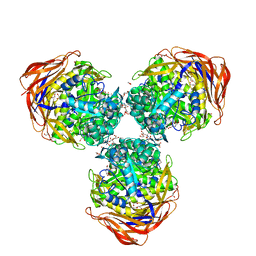 | | Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima ligand free form | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-L-arabinofuranosidase, ... | | Authors: | Im, D.-H, Miyazaki, K, Wakagi, T, Fushinobu, S. | | Deposit date: | 2011-11-02 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Glycoside Hydrolase Family 51 alpha-L-Arabinofuranosidase from Thermotoga maritima
Biosci.Biotechnol.Biochem., 76, 2012
|
|
4ODC
 
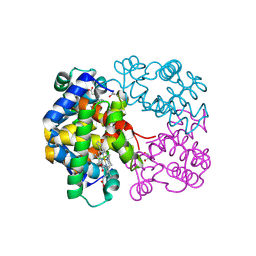 | | Crystal structure of Trematomus bernacchii hemoglobin in a partially cyanided state | | Descriptor: | CYANIDE ION, GLYCEROL, Hemoglobin subunit alpha, ... | | Authors: | Mazzarella, L, Merlino, A, Vitagliano, L, Vergara, A. | | Deposit date: | 2014-01-10 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural modifications induced by the switch from an endogenous bis-histidyl to an exogenous cyanomet hexa-coordination in a tetrameric haemoglobin
RSC ADV, 4, 2014
|
|
3UIV
 
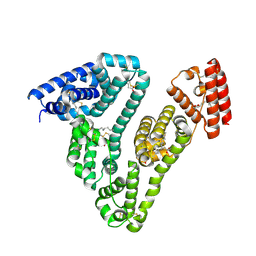 | | Human serum albumin-myristate-amantadine hydrochloride complex | | Descriptor: | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine, MYRISTIC ACID, Serum albumin | | Authors: | Yang, F, Ma, Z, Ma, L, Yang, G. | | Deposit date: | 2011-11-06 | | Release date: | 2012-11-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of the drug-binding specificity of human serum albumin
TO BE PUBLISHED
|
|
4OEG
 
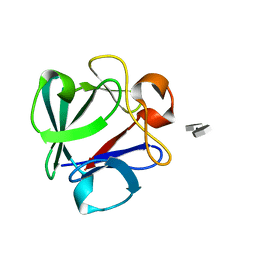 | | Crystal Structure Analysis of FGF2-Disaccharide (S9I2) complex | | Descriptor: | 2-deoxy-3,6-di-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-1-O-methyl-2-O-sulfo-alpha-L-idopyranuronic acid, Fibroblast growth factor 2 | | Authors: | Li, Y.C, Hsiao, C.D. | | Deposit date: | 2014-01-13 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Interactions that influence the binding of synthetic heparan sulfate based disaccharides to fibroblast growth factor-2.
Acs Chem.Biol., 9, 2014
|
|
3UHN
 
 | | HBI (F80Y) deoxy | | Descriptor: | Globin-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ren, Z, Srajer, V, Knapp, J.E, Royer Jr, W.E. | | Deposit date: | 2011-11-03 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cooperative macromolecular device revealed by meta-analysis of static and time-resolved structures.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4ODI
 
 | | 2.6 Angstrom Crystal Structure of Putative Phosphoglycerate Mutase 1 from Toxoplasma gondii | | Descriptor: | Phosphoglycerate mutase PGMII, SODIUM ION | | Authors: | Minasov, G, Ruan, J, Ngo, H, Shuvalova, L, Dubrovska, I, Flores, K, Shanmugam, D, Roos, D, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-01-10 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | CSGID Solves Structures and Identifies Phenotypes for Five Enzymes in Toxoplasma gondii .
Front Cell Infect Microbiol, 8, 2018
|
|
3UHX
 
 | | HBI (N100A) deoxy | | Descriptor: | Globin-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ren, Z, Srajer, V, Knapp, J.E, Royer Jr, W.E. | | Deposit date: | 2011-11-03 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cooperative macromolecular device revealed by meta-analysis of static and time-resolved structures.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2QG6
 
 | |
3UKO
 
 | |
3UKZ
 
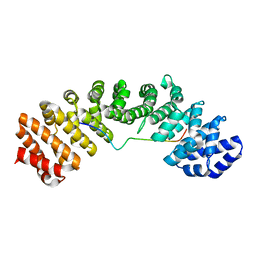 | | Mouse importin alpha: mouse CBP80 cNLS complex | | Descriptor: | Importin subunit alpha-2, Nuclear cap-binding protein subunit 1 | | Authors: | Marfori, M, Forwood, J.K, Lonhienne, T.G, Kobe, B. | | Deposit date: | 2011-11-10 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of High-Affinity Nuclear Localization Signal Interactions with Importin-alpha
Traffic, 13, 2012
|
|
3UIC
 
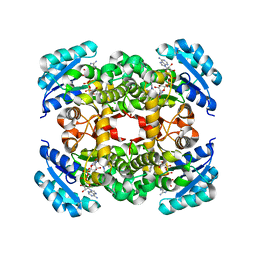 | | Crystal Structure of FabI, an Enoyl Reductase from F. tularensis, in complex with a Novel and Potent Inhibitor | | Descriptor: | 1-(3,4-dichlorobenzyl)-5,6-dimethyl-1H-benzimidazole, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Mehboob, S, Santarsiero, B.D, Truong, K, Johnson, M.E. | | Deposit date: | 2011-11-04 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and enzymatic analyses reveal the binding mode of a novel series of Francisella tularensis enoyl reductase (FabI) inhibitors.
J.Med.Chem., 55, 2012
|
|
3UK7
 
 | |
4OFR
 
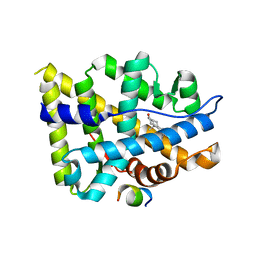 | | Crystal structure of AR-LBD bound with co-regulator peptide | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, co-regulator peptide | | Authors: | Liu, J.S, Hsu, C.L, Wu, W.G. | | Deposit date: | 2014-01-15 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
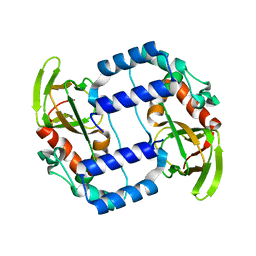
3UKV
 
 | |
3ULI
 
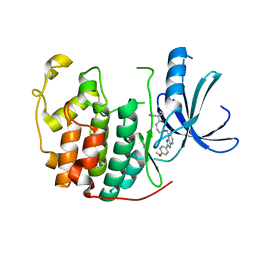 | | Human Cyclin Dependent Kinase 2 (CDK2) bound to azabenzimidazole derivative | | Descriptor: | 1-(aminomethyl)-N-(3-{[6-bromo-2-(4-methoxyphenyl)-3H-imidazo[4,5-b]pyridin-7-yl]amino}propyl)cyclopropanecarboxamide, Cyclin-dependent kinase 2 | | Authors: | Larsen, N.A, Tucker, J.A, Wang, T. | | Deposit date: | 2011-11-10 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of azabenzimidazole derivatives as potent, selective inhibitors of TBK1/IKK epsilon kinases.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4OFU
 
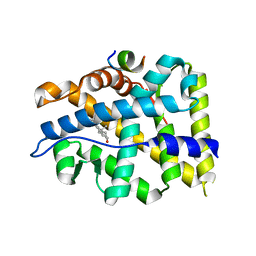 | | Crystal structure of AR-LBD bound with co-regulator peptide | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, co-regulator peptide | | Authors: | Liu, J.S, Hsu, C.L, Wu, W.G. | | Deposit date: | 2014-01-15 | | Release date: | 2014-08-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
3UMA
 
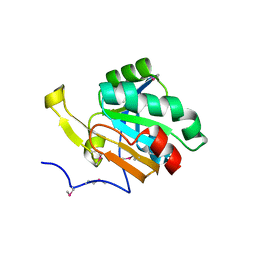 | | Crystal structure of a hypothetical peroxiredoxin protein frm Sinorhizobium meliloti | | Descriptor: | Hypothetical peroxiredoxin protein, SULFATE ION | | Authors: | Eswaramoorthy, S, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-11-12 | | Release date: | 2011-11-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a hypothetical peroxiredoxin protein from Sinorhizobium meliloti
To be Published
|
|
4OOL
 
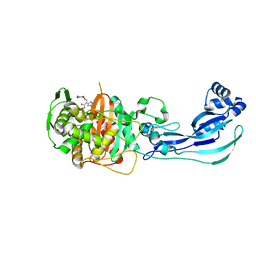 | | Crystal structure of PBP3 in complex with compound 14 ((2E)-2-({[(2S)-2-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(1,5-dihydroxy-4-oxo-1,4-dihydropyridin-2-yl)methoxy]imino}acetyl]amino}-3-oxopropyl]oxy}imino)pentanedioic acid) | | Descriptor: | (2E)-2-({[(2S)-2-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(1,5-dihydroxy-4-oxo-1,4-dihydropyridin-2-yl)methoxy]imino}acetyl]amino}-3-oxopropyl]oxy}imino)pentanedioic acid, Cell division protein FtsI [Peptidoglycan synthetase] | | Authors: | Han, S, Caspers, N, Knafels, J.D. | | Deposit date: | 2014-02-03 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Siderophore receptor-mediated uptake of lactivicin analogues in gram-negative bacteria.
J.Med.Chem., 57, 2014
|
|
3UL1
 
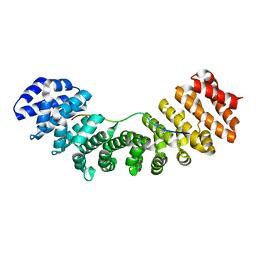 | | Mouse importin alpha: nucleoplasmin cNLS peptide complex | | Descriptor: | Importin subunit alpha-2, Nucleoplasmin | | Authors: | Marfori, M, Forwood, J.K, Lonhienne, T.G, Kobe, B. | | Deposit date: | 2011-11-10 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of High-Affinity Nuclear Localization Signal Interactions with Importin-alpha
Traffic, 13, 2012
|
|
4OFZ
 
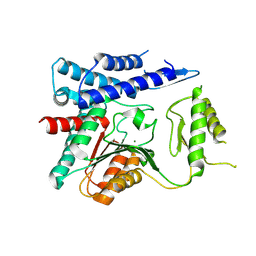 | | Structure of unliganded trehalose-6-phosphate phosphatase from Brugia malayi | | Descriptor: | MAGNESIUM ION, Trehalose-phosphatase | | Authors: | Farelli, J.D, Allen, K.N, Carlow, C.K.S, Dunaway-Mariano, D. | | Deposit date: | 2014-01-15 | | Release date: | 2014-07-16 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Trehalose-6-phosphate Phosphatase from Brugia malayi Reveals Key Design Principles for Anthelmintic Drugs.
Plos Pathog., 10, 2014
|
|
4OP1
 
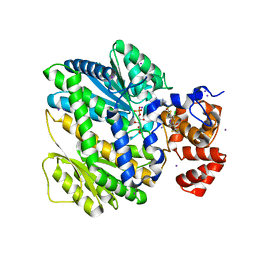 | | GKRP bound to AMG0556 and Sorbitol-6-Phosphate | | Descriptor: | (2S)-2-(6-{5-[(6-aminopyridin-3-yl)sulfonyl]thiophen-2-yl}-5-chloropyridin-3-yl)-1,1,1-trifluoropropan-2-ol, D-SORBITOL-6-PHOSPHATE, GLYCEROL, ... | | Authors: | Jordan, S.R, Chmait, S. | | Deposit date: | 2014-02-04 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Small Molecule Disruptors of the Glucokinase-Glucokinase Regulatory Protein Interaction: 5. A Novel Aryl Sulfone Series, Optimization Through Conformational Analysis.
J.Med.Chem., 58, 2015
|
|
2QA8
 
 | | Crystal Structure of the Estrogen Receptor Alpha Ligand Binding Domain Mutant 537S Complexed with Genistein | | Descriptor: | Estrogen receptor, GENISTEIN, nuclear receptor coactivator 2 | | Authors: | Nettles, K.W, Bruning, J.B, Nowak, J, Sharma, S.K, Hahm, J.B, Shi, Y, Kulp, K, Hochberg, R.B, Zhou, H, Katzenellenbogen, J.A, Katzenellenbogen, B.S, Kim, Y, Joachmiak, A, Greene, G.L. | | Deposit date: | 2007-06-14 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | NFkappaB selectivity of estrogen receptor ligands revealed by comparative crystallographic analyses
Nat.Chem.Biol., 4, 2008
|
|
3UN3
 
 | | phosphopentomutase T85Q variant soaked with glucose 1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-alpha-D-glucopyranose, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Iverson, T.M, Birmingham, W.R, Panosian, T.D, Nannemann, D.P, Bachmann, B.O. | | Deposit date: | 2011-11-15 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Differences between a Mutase and a Phosphatase: Investigations of the Activation Step in Bacillus cereus Phosphopentomutase.
Biochemistry, 51, 2012
|
|
4OPH
 
 | | X-ray structure of full-length H6N6 NS1 | | Descriptor: | Nonstructural protein 1 | | Authors: | Carrillo, B, Choi, J.M, Bornholdt, Z.A, Sankaran, S, Rice, A.P, Prasad, B.V.V. | | Deposit date: | 2014-02-05 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.158 Å) | | Cite: | The Influenza A Virus Protein NS1 Displays Structural Polymorphism.
J.Virol., 88, 2014
|
|
3UBX
 
 | | Crystal structure of the mouse CD1d-C20:2-aGalCer-L363 mAb Fab complex | | Descriptor: | (11Z,14Z)-N-[(2S,3S,4R)-1-(alpha-D-galactopyranosyloxy)-3,4-dihydroxyoctadecan-2-yl]icosa-11,14-dienamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yu, E.D, Zajonc, D.M. | | Deposit date: | 2011-10-25 | | Release date: | 2011-11-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the recognition of C20:2-alpha GalCer by the invariant natural killer T cell receptor-like antibody L363.
J.Biol.Chem., 287, 2012
|
|
