7K0Y
 
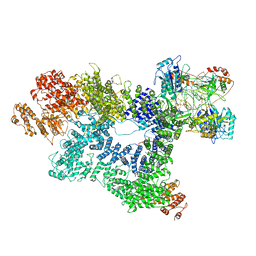 | | Cryo-EM structure of activated-form DNA-PK (complex VI) | | Descriptor: | DNA (5'-D(*AP*AP*GP*CP*AP*GP*TP*AP*GP*AP*GP*CP*A)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*CP*TP*CP*TP*AP*CP*TP*GP*CP*TP*TP*CP*GP*AP*TP*AP*TP*CP*G)-3'), DNA-dependent protein kinase catalytic subunit, ... | | Authors: | Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2020-09-06 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of an activated DNA-PK and its implications for NHEJ.
Mol.Cell, 81, 2021
|
|
7K11
 
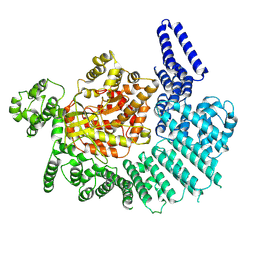 | |
7K10
 
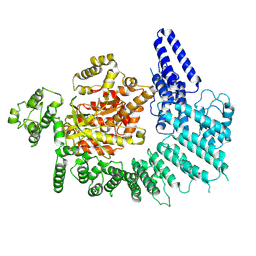 | |
6F3L
 
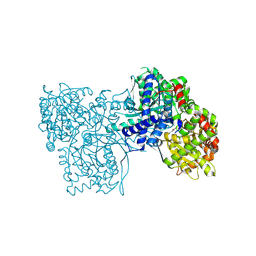 | | The crystal structure of Glycogen Phosphorylase in complex with 10b | | Descriptor: | 6-[5-[(2~{S},3~{R},4~{R},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]-1~{H}-1,2,4-triazol-3-yl]naphthalene-2-carboxylic acid, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Barkas, T.A, Stravodimos, G.A, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2017-11-28 | | Release date: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A multidisciplinary study of 3-( beta-d-glucopyranosyl)-5-substituted-1,2,4-triazole derivatives as glycogen phosphorylase inhibitors: Computation, synthesis, crystallography and kinetics reveal new potent inhibitors.
Eur J Med Chem, 147, 2018
|
|
7DNT
 
 | | mRNA-decapping enzyme g5Rp | | Descriptor: | mRNA-decapping protein g5R | | Authors: | Yang, Y, Chen, C, Li, L, Li, X.H, Su, D. | | Deposit date: | 2020-12-10 | | Release date: | 2022-03-09 | | Last modified: | 2022-12-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insight into Molecular Inhibitory Mechanism of InsP 6 on African Swine Fever Virus mRNA-Decapping Enzyme g5Rp.
J.Virol., 96, 2022
|
|
6FM3
 
 | | Deoxyguanylosuccinate synthase (DgsS) structure with ADP at 1.9 Angstrom resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Adenylosuccinate synthetase, CHLORIDE ION | | Authors: | Sleiman, D, Loc'h, J, Haouz, A, Kaminski, P.A. | | Deposit date: | 2018-01-30 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Deoxyguanylosuccinate synthase (DgsS) quaternary structure with ATP0, dGMP, Magnesium at 2.3 Angstrom resolution
To Be Published
|
|
6VZ4
 
 | |
5K8H
 
 | |
7O6P
 
 | |
6NUI
 
 | | Human Guanylate Kinase | | Descriptor: | Guanylate kinase | | Authors: | Sabo, T.M, Khan, N, Ban, D, Trigo-Mourino, P, Carneiro, M.G, Trent, J.O, Konrad, M, Lee, D. | | Deposit date: | 2019-02-01 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and functional investigation of human guanylate kinase reveals allosteric networking and a crucial role for the enzyme in cancer.
J.Biol.Chem., 294, 2019
|
|
7ODJ
 
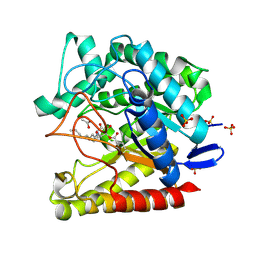 | | Exo-mannosidase from Cellvibrio mixtus bound to N-alkyl mannocyclophellitol aziridine | | Descriptor: | (1R,2S,3R,4S,5R,6R)-5-(8-azidooctylamino)-6-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, ACETATE ION, Man5A, ... | | Authors: | McGregor, N.G.S, Davies, G.J. | | Deposit date: | 2021-04-29 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Synthesis of broad-specificity activity-based probes for exo -beta-mannosidases.
Org.Biomol.Chem., 20, 2022
|
|
7T5Q
 
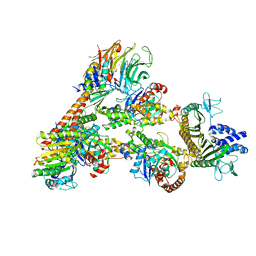 | | Cryo-EM Structure of a Transition State of Arp2/3 Complex Activation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Rebowski, G, van Eeuwen, T, Boczkowska, M, Dominguez, R. | | Deposit date: | 2021-12-13 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Transition State of Arp2/3 Complex Activation by Actin-Bound Dimeric Nucleation-Promoting Factor.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7NXI
 
 | | PAF-D19S in 50 v/v % DMSO-water solution | | Descriptor: | Pc24g00380 protein | | Authors: | Czajlik, A, Batta, G. | | Deposit date: | 2021-03-18 | | Release date: | 2022-03-30 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | DMSO-Induced Unfolding of the Antifungal Disulfide Protein PAF and Its Inactive Variant: A Combined NMR and DSC Study.
Int J Mol Sci, 24, 2023
|
|
7O3D
 
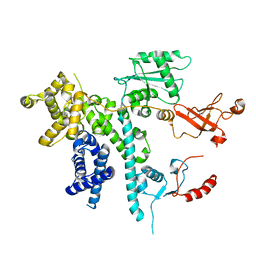 | | Cooperation between the intrinsically disordered and ordered regions of Spt6 regulates nucleosome and Pol II CTD binding, and nucleosome assembly | | Descriptor: | Transcription elongation factor SPT6 | | Authors: | Kasiliauskaite, A, Kubicek, K, Klumpler, T, Zanova, M, Zapletal, D, Novacek, J, Stefl, R. | | Deposit date: | 2021-04-01 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Cooperation between intrinsically disordered and ordered regions of Spt6 regulates nucleosome and Pol II CTD binding, and nucleosome assembly.
Nucleic Acids Res., 50, 2022
|
|
7O6B
 
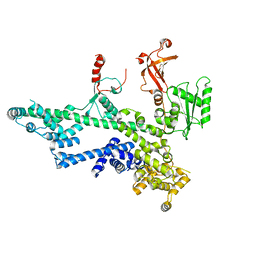 | | Cooperation between the intrinsically disordered and ordered regions of Spt6 regulates nucleosome and Pol II CTD binding, and nucleosome assembly | | Descriptor: | Transcription elongation factor SPT6 | | Authors: | Kasiliauskaite, A, Kubicek, K, Klumpler, T, Zanova, M, Zapletal, D, Novacek, J, Stefl, R. | | Deposit date: | 2021-04-09 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Cooperation between intrinsically disordered and ordered regions of Spt6 regulates nucleosome and Pol II CTD binding, and nucleosome assembly.
Nucleic Acids Res., 50, 2022
|
|
7L5X
 
 | | p97-R155H mutant dodecamer II | | Descriptor: | Transitional endoplasmic reticulum ATPase | | Authors: | Nandi, P, Li, S, Coulmbres, R.C.A, Wang, F, Williams, D.R, Malyutin, A.G, Poh, Y.-P, Chou, T.-F, Chiu, P.-L. | | Deposit date: | 2020-12-23 | | Release date: | 2021-08-04 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structural and Functional Analysis of Disease-Linked p97 ATPase Mutant Complexes.
Int J Mol Sci, 22, 2021
|
|
7QP6
 
 | | Structure of the human 48S initiation complex in open state (h48S AUG open) | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Yi, S.-H, Petrychenko, V, Schliep, J.E, Goyal, A, Linden, A, Chari, A, Urlaub, H, Stark, H, Rodnina, M.V, Adio, S, Fischer, N. | | Deposit date: | 2022-01-03 | | Release date: | 2022-05-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Conformational rearrangements upon start codon recognition in human 48S translation initiation complex.
Nucleic Acids Res., 50, 2022
|
|
7QP7
 
 | | Structure of the human 48S initiation complex in closed state (h48S AUG closed) | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Yi, S.-H, Petrychenko, V, Schliep, J.E, Goyal, A, Linden, A, Chari, A, Urlaub, H, Stark, H, Rodnina, M.V, Adio, S, Fischer, N. | | Deposit date: | 2022-01-03 | | Release date: | 2022-05-11 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Conformational rearrangements upon start codon recognition in human 48S translation initiation complex.
Nucleic Acids Res., 50, 2022
|
|
6R0C
 
 | | Human-D02 Nucleosome Core Particle with biotin-streptavidin label | | Descriptor: | DNA (142-MER), Histone H2A type 1, Histone H2B type 1-C/E/F/G/I, ... | | Authors: | Pye, V.E, Wilson, M.D, Cherepanov, P, Costa, A. | | Deposit date: | 2019-03-12 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Retroviral integration into nucleosomes through DNA looping and sliding along the histone octamer.
Nat Commun, 10, 2019
|
|
7ODE
 
 | | E. coli 50S ribosome LiCl core particle | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Larsson, D.S.D, Selmer, M. | | Deposit date: | 2021-04-29 | | Release date: | 2022-06-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural Consequences of Deproteinating the 50S Ribosome.
Biomolecules, 12, 2022
|
|
7ZNJ
 
 | | Structure of an ALYREF-exon junction complex hexamer | | Descriptor: | Eukaryotic initiation factor 4A-III, N-terminally processed, MAGNESIUM ION, ... | | Authors: | Pacheco-Fiallos, F.B, Vorlaender, M.K, Plaschka, C. | | Deposit date: | 2022-04-21 | | Release date: | 2023-04-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | mRNA recognition and packaging by the human transcription-export complex.
Nature, 616, 2023
|
|
7LAR
 
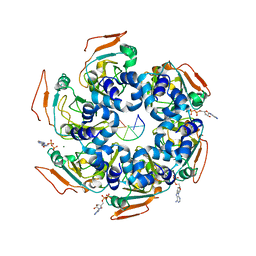 | | Cryo-EM structure of PCV2 Replicase bound to ssDNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent helicase Rep, DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Khayat, R. | | Deposit date: | 2021-01-06 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Mechanism of DNA Interaction and Translocation by the Replicase of a Circular Rep-Encoding Single-Stranded DNA Virus.
Mbio, 12, 2021
|
|
7LAS
 
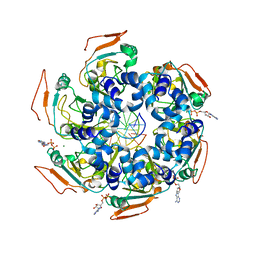 | | Cryo-EM structure of PCV2 Replicase bound to ssDNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent helicase Rep, DNA (5'-D(P*GP*AP*TP*CP*GP*AP*TP*CP*GP*A)-3'), ... | | Authors: | Khayat, R. | | Deposit date: | 2021-01-06 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Mechanism of DNA Interaction and Translocation by the Replicase of a Circular Rep-Encoding Single-Stranded DNA Virus.
Mbio, 12, 2021
|
|
7ZI4
 
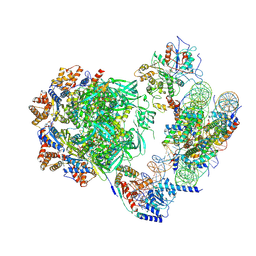 | | Cryo-EM structure of the human INO80 complex bound to a WT nucleosome | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-related protein 5, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Vance, N.R, Ayala, R, Willhoft, O, Tvardovskiy, A, McCormack, E.A, Bartke, T, Zhang, X, Wigley, D.B. | | Deposit date: | 2022-04-07 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the human INO80 complex bound to a WT nucleosome
To Be Published
|
|
5IQ5
 
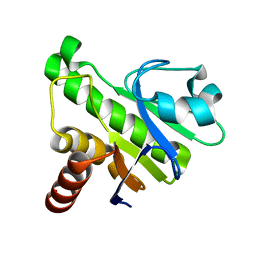 | | NMR solution structure of Mayaro virus macro domain | | Descriptor: | Macro domain | | Authors: | Melekis, E, Tsika, A.C, Bentrop, D, Papageorgiou, N, Coutard, B, Spyroulias, G.A. | | Deposit date: | 2016-03-10 | | Release date: | 2017-12-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Deciphering the Nucleotide and RNA Binding Selectivity of the Mayaro Virus Macro Domain.
J.Mol.Biol., 431, 2019
|
|
